Liren Jiang
Artificial intelligence for diagnosing and predicting survival of patients with renal cell carcinoma: Retrospective multi-center study
Jan 12, 2023



Abstract:Background: Clear cell renal cell carcinoma (ccRCC) is the most common renal-related tumor with high heterogeneity. There is still an urgent need for novel diagnostic and prognostic biomarkers for ccRCC. Methods: We proposed a weakly-supervised deep learning strategy using conventional histology of 1752 whole slide images from multiple centers. Our study was demonstrated through internal cross-validation and external validations for the deep learning-based models. Results: Automatic diagnosis for ccRCC through intelligent subtyping of renal cell carcinoma was proved in this study. Our graderisk achieved aera the curve (AUC) of 0.840 (95% confidence interval: 0.805-0.871) in the TCGA cohort, 0.840 (0.805-0.871) in the General cohort, and 0.840 (0.805-0.871) in the CPTAC cohort for the recognition of high-grade tumor. The OSrisk for the prediction of 5-year survival status achieved AUC of 0.784 (0.746-0.819) in the TCGA cohort, which was further verified in the independent General cohort and the CPTAC cohort, with AUC of 0.774 (0.723-0.820) and 0.702 (0.632-0.765), respectively. Cox regression analysis indicated that graderisk, OSrisk, tumor grade, and tumor stage were found to be independent prognostic factors, which were further incorporated into the competing-risk nomogram (CRN). Kaplan-Meier survival analyses further illustrated that our CRN could significantly distinguish patients with high survival risk, with hazard ratio of 5.664 (3.893-8.239, p < 0.0001) in the TCGA cohort, 35.740 (5.889-216.900, p < 0.0001) in the General cohort and 6.107 (1.815 to 20.540, p < 0.0001) in the CPTAC cohort. Comparison analyses conformed that our CRN outperformed current prognosis indicators in the prediction of survival status, with higher concordance index for clinical prognosis.
Large-scale Gastric Cancer Screening and Localization Using Multi-task Deep Neural Network
Oct 12, 2019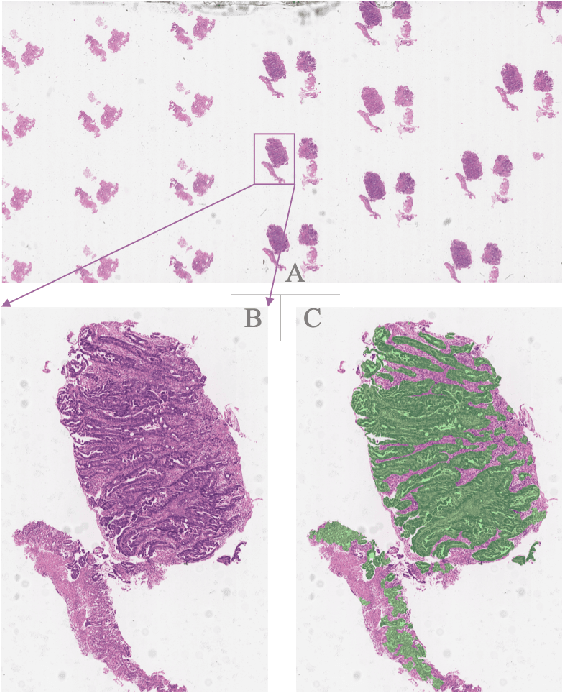
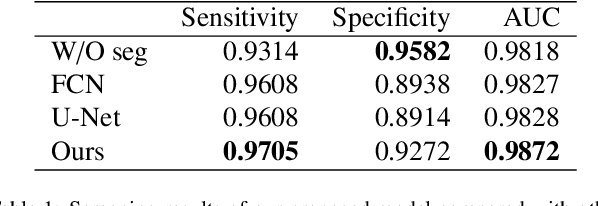
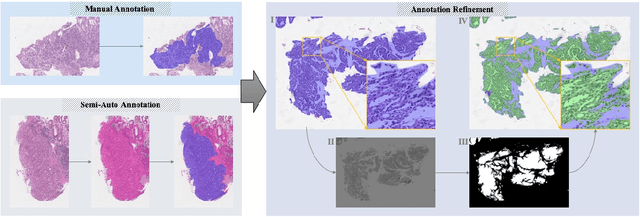
Abstract:Gastric cancer is one of the most common cancers, which ranks third among the leading causes of cancer death. Biopsy of gastric mucosal is a standard procedure in gastric cancer screening test. However, manual pathological inspection is labor-intensive and time-consuming. Besides, it is challenging for an automated algorithm to locate the small lesion regions in the gigapixel whole-slide image and make the decision correctly. To tackle these issues, we collected large-scale whole-slide image dataset with detailed lesion region annotation and designed a whole-slide image analyzing framework consisting of 3 networks which could not only determine the screen result but also present the suspicious areas to the pathologist for reference. Experiments demonstrated that our proposed framework achieves sensitivity of 97.05% and specificity of 92.72% in screening task and Dice coefficient of 0.8331 in segmentation task. Furthermore, we tested our best model in real-world scenario on 10, 316 whole-slide images collected from 4 medical centers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge