Wenhua Zhang
MMM-RS: A Multi-modal, Multi-GSD, Multi-scene Remote Sensing Dataset and Benchmark for Text-to-Image Generation
Oct 26, 2024



Abstract:Recently, the diffusion-based generative paradigm has achieved impressive general image generation capabilities with text prompts due to its accurate distribution modeling and stable training process. However, generating diverse remote sensing (RS) images that are tremendously different from general images in terms of scale and perspective remains a formidable challenge due to the lack of a comprehensive remote sensing image generation dataset with various modalities, ground sample distances (GSD), and scenes. In this paper, we propose a Multi-modal, Multi-GSD, Multi-scene Remote Sensing (MMM-RS) dataset and benchmark for text-to-image generation in diverse remote sensing scenarios. Specifically, we first collect nine publicly available RS datasets and conduct standardization for all samples. To bridge RS images to textual semantic information, we utilize a large-scale pretrained vision-language model to automatically output text prompts and perform hand-crafted rectification, resulting in information-rich text-image pairs (including multi-modal images). In particular, we design some methods to obtain the images with different GSD and various environments (e.g., low-light, foggy) in a single sample. With extensive manual screening and refining annotations, we ultimately obtain a MMM-RS dataset that comprises approximately 2.1 million text-image pairs. Extensive experimental results verify that our proposed MMM-RS dataset allows off-the-shelf diffusion models to generate diverse RS images across various modalities, scenes, weather conditions, and GSD. The dataset is available at https://github.com/ljl5261/MMM-RS.
Automated Evaluation of Large Vision-Language Models on Self-driving Corner Cases
Apr 16, 2024



Abstract:Large Vision-Language Models (LVLMs), due to the remarkable visual reasoning ability to understand images and videos, have received widespread attention in the autonomous driving domain, which significantly advances the development of interpretable end-to-end autonomous driving. However, current evaluations of LVLMs primarily focus on the multi-faceted capabilities in common scenarios, lacking quantifiable and automated assessment in autonomous driving contexts, let alone severe road corner cases that even the state-of-the-art autonomous driving perception systems struggle to handle. In this paper, we propose CODA-LM, a novel vision-language benchmark for self-driving, which provides the first automatic and quantitative evaluation of LVLMs for interpretable autonomous driving including general perception, regional perception, and driving suggestions. CODA-LM utilizes the texts to describe the road images, exploiting powerful text-only large language models (LLMs) without image inputs to assess the capabilities of LVLMs in autonomous driving scenarios, which reveals stronger alignment with human preferences than LVLM judges. Experiments demonstrate that even the closed-sourced commercial LVLMs like GPT-4V cannot deal with road corner cases well, suggesting that we are still far from a strong LVLM-powered intelligent driving agent, and we hope our CODA-LM can become the catalyst to promote future development.
Hybrid-SORT: Weak Cues Matter for Online Multi-Object Tracking
Aug 01, 2023Abstract:Multi-Object Tracking (MOT) aims to detect and associate all desired objects across frames. Most methods accomplish the task by explicitly or implicitly leveraging strong cues (i.e., spatial and appearance information), which exhibit powerful instance-level discrimination. However, when object occlusion and clustering occur, both spatial and appearance information will become ambiguous simultaneously due to the high overlap between objects. In this paper, we demonstrate that this long-standing challenge in MOT can be efficiently and effectively resolved by incorporating weak cues to compensate for strong cues. Along with velocity direction, we introduce the confidence state and height state as potential weak cues. With superior performance, our method still maintains Simple, Online and Real-Time (SORT) characteristics. Furthermore, our method shows strong generalization for diverse trackers and scenarios in a plug-and-play and training-free manner. Significant and consistent improvements are observed when applying our method to 5 different representative trackers. Further, by leveraging both strong and weak cues, our method Hybrid-SORT achieves superior performance on diverse benchmarks, including MOT17, MOT20, and especially DanceTrack where interaction and occlusion are frequent and severe. The code and models are available at https://github.com/ymzis69/HybirdSORT.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
dual unet:a novel siamese network for change detection with cascade differential fusion
Aug 12, 2022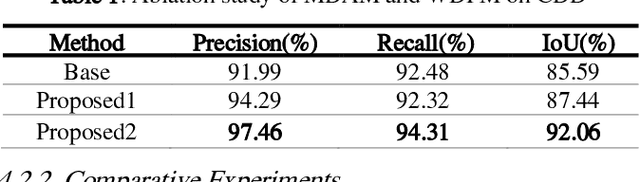
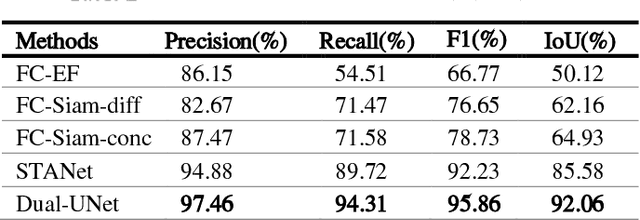
Abstract:Change detection (CD) of remote sensing images is to detect the change region by analyzing the difference between two bitemporal images. It is extensively used in land resource planning, natural hazards monitoring and other fields. In our study, we propose a novel Siamese neural network for change detection task, namely Dual-UNet. In contrast to previous individually encoded the bitemporal images, we design an encoder differential-attention module to focus on the spatial difference relationships of pixels. In order to improve the generalization of networks, it computes the attention weights between any pixels between bitemporal images and uses them to engender more discriminating features. In order to improve the feature fusion and avoid gradient vanishing, multi-scale weighted variance map fusion strategy is proposed in the decoding stage. Experiments demonstrate that the proposed approach consistently outperforms the most advanced methods on popular seasonal change detection datasets.
A Dual-fusion Semantic Segmentation Framework With GAN For SAR Images
Jun 02, 2022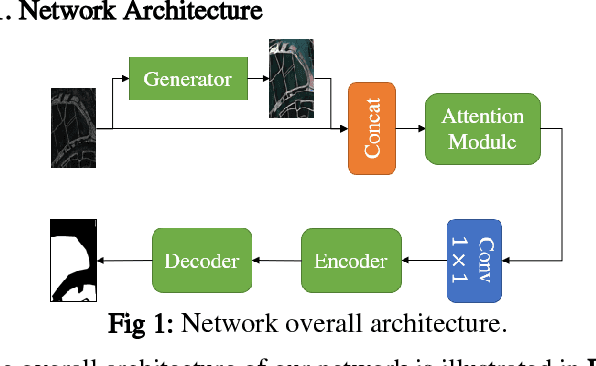
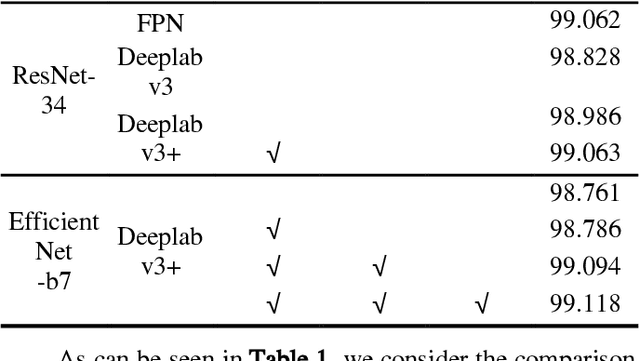
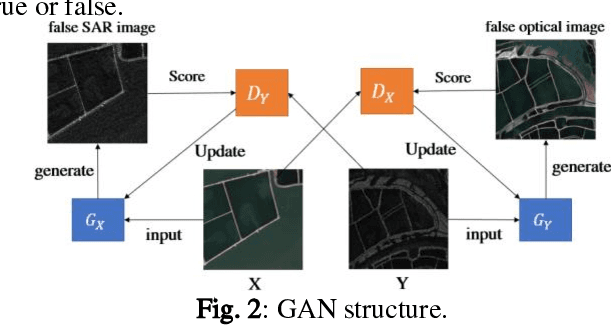
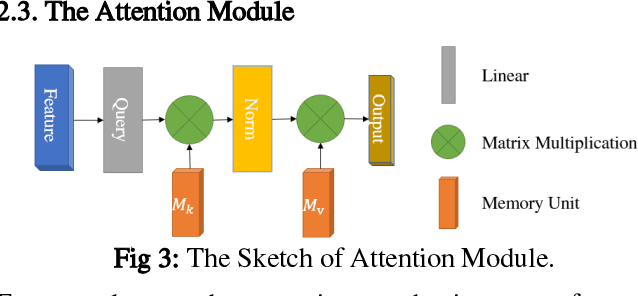
Abstract:Deep learning based semantic segmentation is one of the popular methods in remote sensing image segmentation. In this paper, a network based on the widely used encoderdecoder architecture is proposed to accomplish the synthetic aperture radar (SAR) images segmentation. With the better representation capability of optical images, we propose to enrich SAR images with generated optical images via the generative adversative network (GAN) trained by numerous SAR and optical images. These optical images can be used as expansions of original SAR images, thus ensuring robust result of segmentation. Then the optical images generated by the GAN are stitched together with the corresponding real images. An attention module following the stitched data is used to strengthen the representation of the objects. Experiments indicate that our method is efficient compared to other commonly used methods
AugHover-Net: Augmenting Hover-net for Nucleus Segmentation and Classification
Apr 02, 2022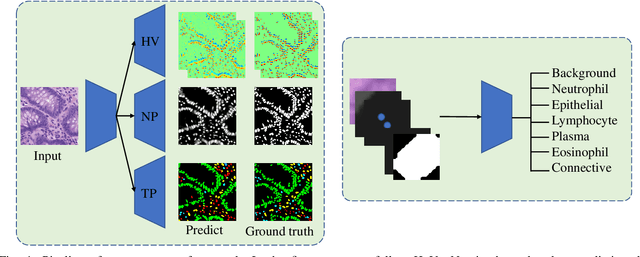
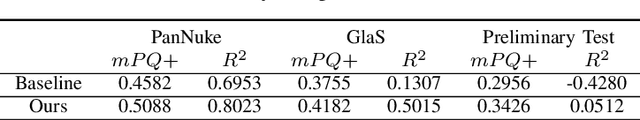
Abstract:Nuclei segmentation and classification have been a challenge in digital pathology due to the specific domain characteristics. First, annotating a large-scale dataset is quite consuming. It requires specific domain knowledge and large efforts. Second, some nuclei are clustered together and hard to segment from each other. Third, the classes are often extremely unbalanced. As in Lizard, the number of epithelial nuclei is around 67 times larger than the number of eosinophil nuclei. Fourth, the nuclei often exhibit high inter-class similarity and intra-class variability. Connective nuclei may look very different from each other while some of them share a similar shape with the epithelial ones. Last but not least, pathological patches may have very different color distributions among different datasets. Thus, a large-scale generally annotated dataset and a specially-designed algorithm are needed to solve this problem. The CoNIC challenge aims to promote the automatic segmentation and classification task and requires researchers to develop algorithms that perform segmentation, classification, and counting of 6 different types of nuclei with the large-scale annotated dataset: Lizard. Due to the 60-minute time limit, the algorithm has to be simple and quick. In this paper, we briefly describe the final method we used in the CoNIC challenge. Our algorithm is based on Hover-Net and we added several modifications to it to improve its performance.
Hierarchical Feature-Aware Tracking
Oct 18, 2019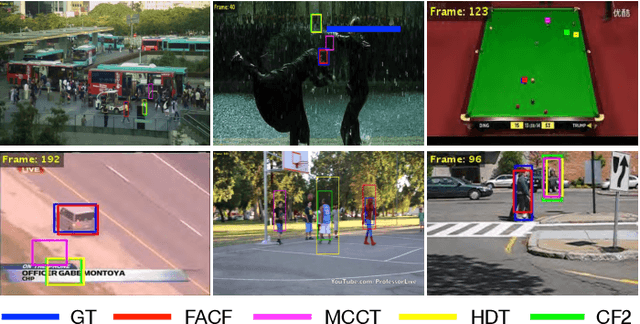
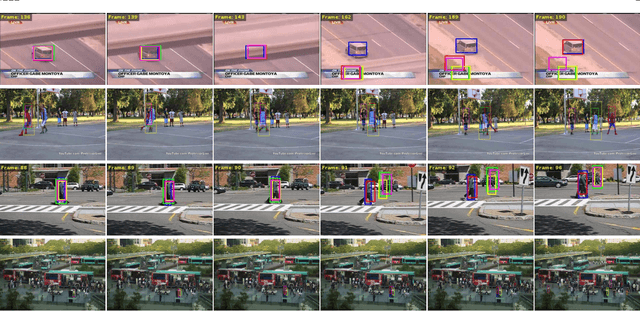
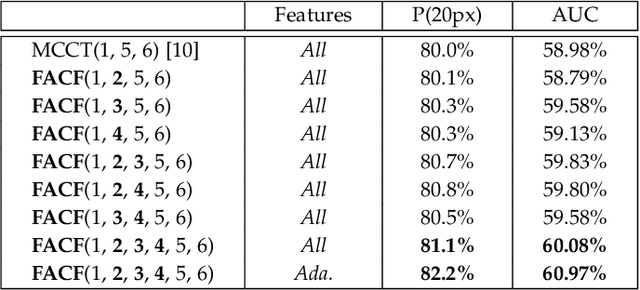
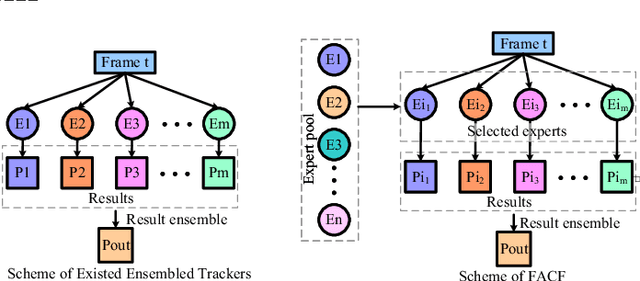
Abstract:In this paper, we propose a hierarchical feature-aware tracking framework for efficient visual tracking. Recent years, ensembled trackers which combine multiple component trackers have achieved impressive performance. In ensembled trackers, the decision of results is usually a post-event process, i.e., tracking result for each tracker is first obtained and then the suitable one is selected according to result ensemble. In this paper, we propose a pre-event method. We construct an expert pool with each expert being one set of features. For each frame, several experts are first selected in the pool according to their past performance and then they are used to predict the object. The selection rate of each expert in the pool is then updated and tracking result is obtained according to result ensemble. We propose a novel pre-known expert-adaptive selection strategy. Since the process is more efficient, more experts can be constructed by fusing more types of features which leads to more robustness. Moreover, with the novel expert selection strategy, overfitting caused by fixed experts for each frame can be mitigated. Experiments on several public available datasets demonstrate the superiority of the proposed method and its state-of-the-art performance among ensembled trackers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge