Uwe Schmidt
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Nuclei instance segmentation and classification in histopathology images with StarDist
Mar 24, 2022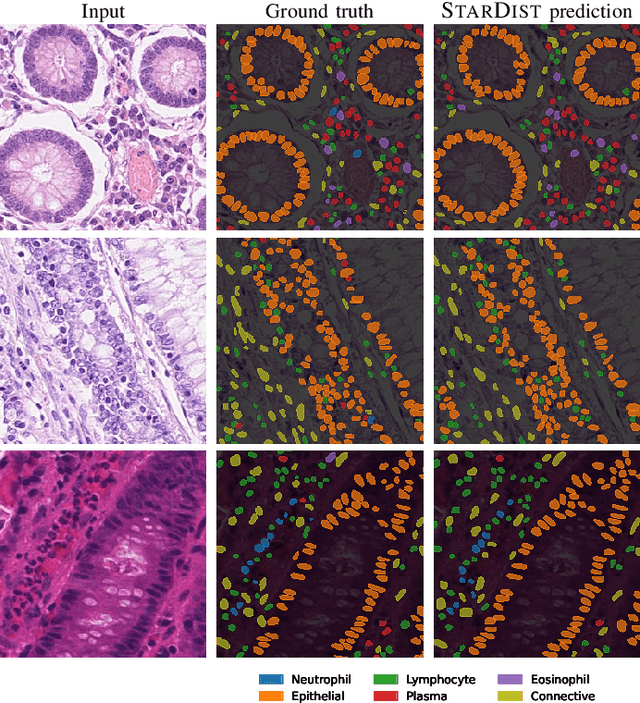
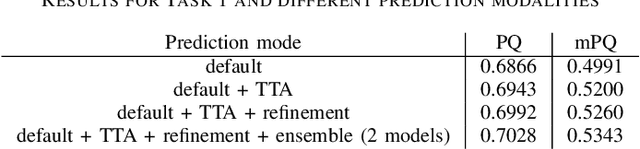
Abstract:Instance segmentation and classification of nuclei is an important task in computational pathology. We show that StarDist, a deep learning based nuclei segmentation method originally developed for fluorescence microscopy, can be extended and successfully applied to histopathology images. This is substantiated by conducting experiments on the Lizard dataset, and through entering the Colon Nuclei Identification and Counting (CoNIC) challenge 2022. At the end of the preliminary test phase of CoNIC, our approach ranked first on the leaderboard for the segmentation and classification task.
Fast and accurate aberration estimation from 3D bead images using convolutional neural networks
Jun 02, 2020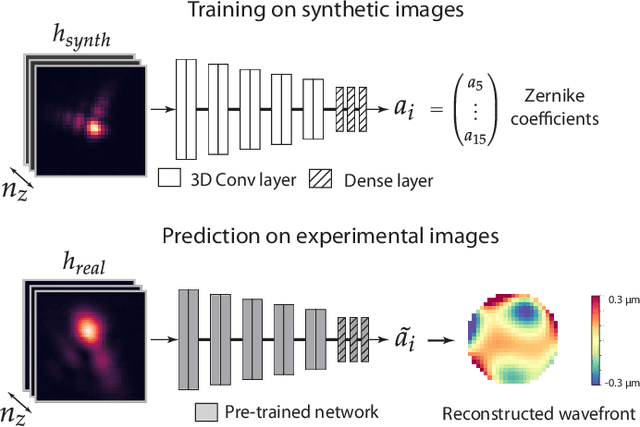

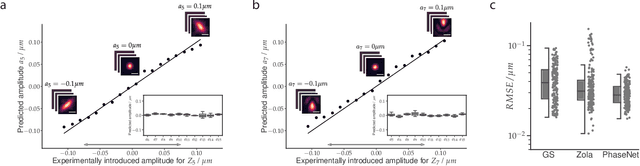
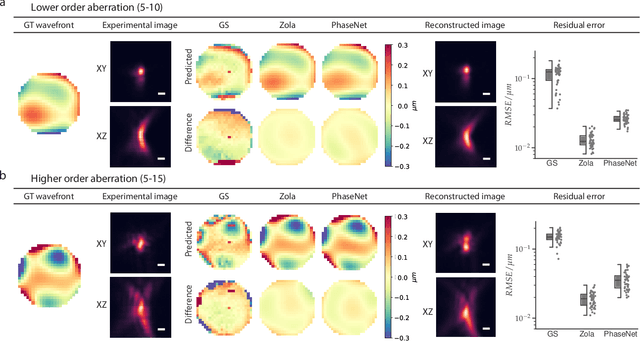
Abstract:Estimating optical aberrations from volumetric intensity images is a key step in sensorless adaptive optics for microscopy. Here we describe a method (PHASENET) for fast and accurate aberration measurement from experimentally acquired 3D bead images using convolutional neural networks. Importantly, we show that networks trained only on synthetically generated data can successfully predict aberrations from experimental images. We demonstrate our approach on two data sets acquired with different microscopy modalities and find that PHASENET yields results better than or comparable to classical methods while being orders of magnitude faster. We furthermore show that the number of focal planes required for satisfactory prediction is related to different symmetry groups of Zernike modes. PHASENET is freely available as open-source software in Python.
An interpretable automated detection system for FISH-based HER2 oncogene amplification testing in histo-pathological routine images of breast and gastric cancer diagnostics
May 25, 2020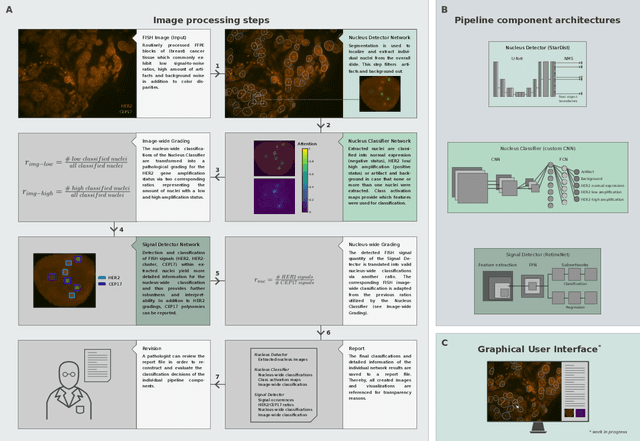
Abstract:Histo-pathological diagnostics are an inherent part of the everyday work but are particularly laborious and associated with time-consuming manual analysis of image data. In order to cope with the increasing diagnostic case numbers due to the current growth and demographic change of the global population and the progress in personalized medicine, pathologists ask for assistance. Profiting from digital pathology and the use of artificial intelligence, individual solutions can be offered (e.g. detect labeled cancer tissue sections). The testing of the human epidermal growth factor receptor 2 (HER2) oncogene amplification status via fluorescence in situ hybridization (FISH) is recommended for breast and gastric cancer diagnostics and is regularly performed at clinics. Here, we develop an interpretable, deep learning (DL)-based pipeline which automates the evaluation of FISH images with respect to HER2 gene amplification testing. It mimics the pathological assessment and relies on the detection and localization of interphase nuclei based on instance segmentation networks. Furthermore, it localizes and classifies fluorescence signals within each nucleus with the help of image classification and object detection convolutional neural networks (CNNs). Finally, the pipeline classifies the whole image regarding its HER2 amplification status. The visualization of pixels on which the networks' decision occurs, complements an essential part to enable interpretability by pathologists.
Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy
Aug 09, 2019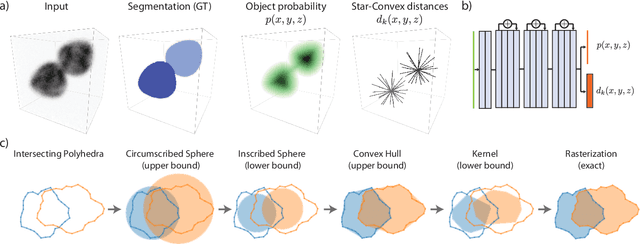
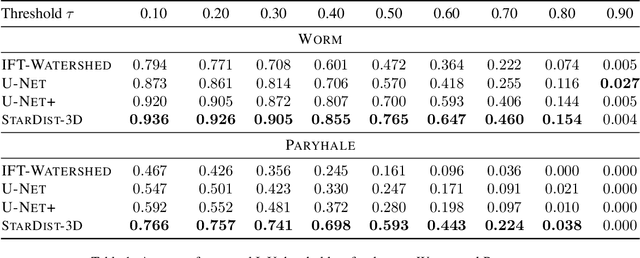
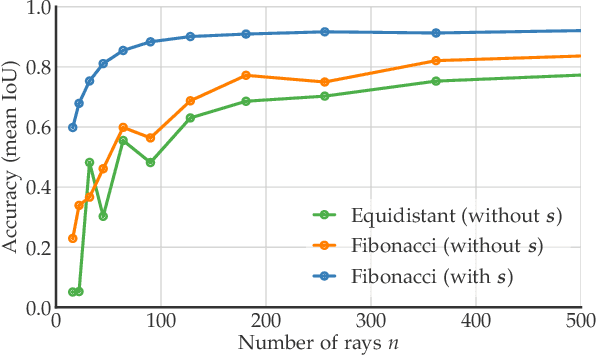
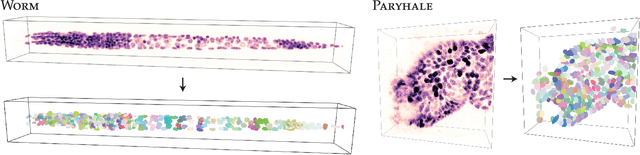
Abstract:Accurate detection and segmentation of cell nuclei in volumetric (3D) fluorescence microscopy datasets is an important step in many biomedical research projects. Although many automated methods for these tasks exist, they often struggle for images with low signal-to-noise ratios and/or dense packing of nuclei. It was recently shown for 2D microscopy images that these issues can be alleviated by training a neural network to directly predict a suitable shape representation (star-convex polygon) for cell nuclei. In this paper, we adopt and extend this approach to 3D volumes by using star-convex polyhedra to represent cell nuclei and similar shapes. To that end, we overcome the challenges of 1) finding parameter-efficient star-convex polyhedra representations that can faithfully describe cell nuclei shapes, 2) adapting to anisotropic voxel sizes often found in fluorescence microscopy datasets, and 3) efficiently computing intersections between pairs of star-convex polyhedra (required for non-maximum suppression). Although our approach is quite general, since star-convex polyhedra subsume common shapes like bounding boxes and spheres as special cases, our focus is on accurate detection and segmentation of cell nuclei. That that end, we demonstrate on two challenging datasets that our approach (StarDist-3D) leads to superior results when compared to classical and deep-learning based methods.
Cell Detection with Star-convex Polygons
Jun 09, 2018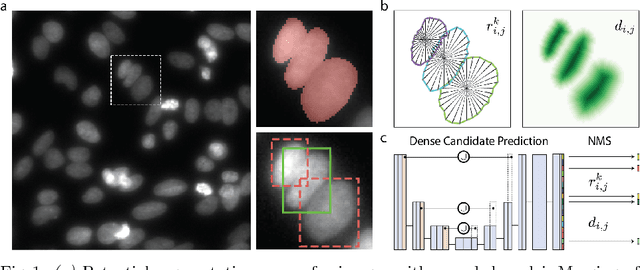
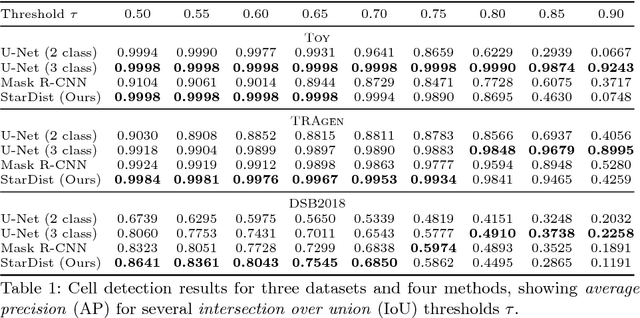
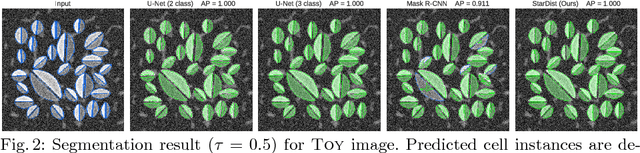
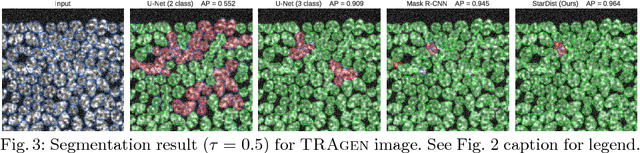
Abstract:Automatic detection and segmentation of cells and nuclei in microscopy images is important for many biological applications. Recent successful learning-based approaches include per-pixel cell segmentation with subsequent pixel grouping, or localization of bounding boxes with subsequent shape refinement. In situations of crowded cells, these can be prone to segmentation errors, such as falsely merging bordering cells or suppressing valid cell instances due to the poor approximation with bounding boxes. To overcome these issues, we propose to localize cell nuclei via star-convex polygons, which are a much better shape representation as compared to bounding boxes and thus do not need shape refinement. To that end, we train a convolutional neural network that predicts for every pixel a polygon for the cell instance at that position. We demonstrate the merits of our approach on two synthetic datasets and one challenging dataset of diverse fluorescence microscopy images.
Cascades of Regression Tree Fields for Image Restoration
Nov 21, 2014
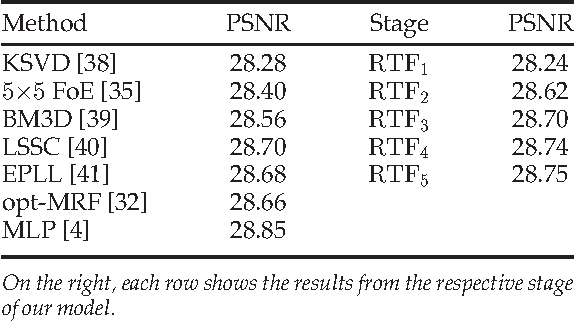
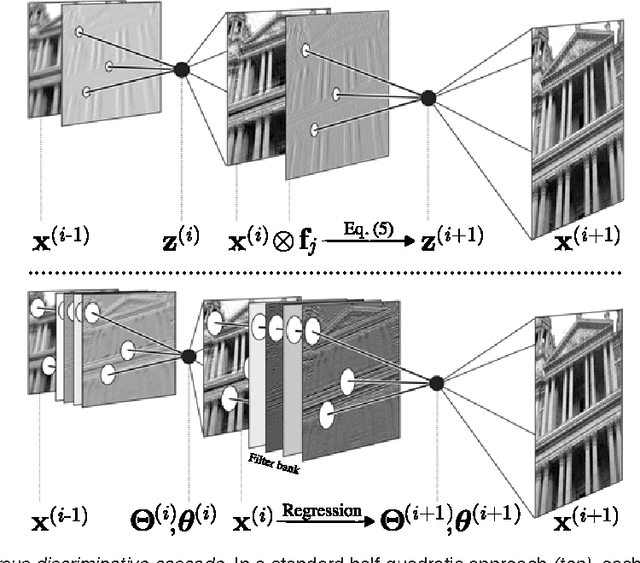
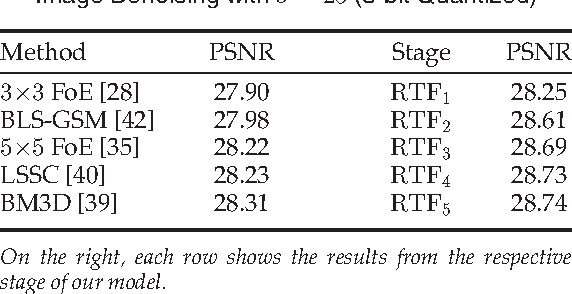
Abstract:Conditional random fields (CRFs) are popular discriminative models for computer vision and have been successfully applied in the domain of image restoration, especially to image denoising. For image deblurring, however, discriminative approaches have been mostly lacking. We posit two reasons for this: First, the blur kernel is often only known at test time, requiring any discriminative approach to cope with considerable variability. Second, given this variability it is quite difficult to construct suitable features for discriminative prediction. To address these challenges we first show a connection between common half-quadratic inference for generative image priors and Gaussian CRFs. Based on this analysis, we then propose a cascade model for image restoration that consists of a Gaussian CRF at each stage. Each stage of our cascade is semi-parametric, i.e. it depends on the instance-specific parameters of the restoration problem, such as the blur kernel. We train our model by loss minimization with synthetically generated training data. Our experiments show that when applied to non-blind image deblurring, the proposed approach is efficient and yields state-of-the-art restoration quality on images corrupted with synthetic and real blur. Moreover, we demonstrate its suitability for image denoising, where we achieve competitive results for grayscale and color images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge