Liang Dong
refer to the report for detailed contributions
SFMP: Fine-Grained, Hardware-Friendly and Search-Free Mixed-Precision Quantization for Large Language Models
Feb 01, 2026Abstract:Mixed-precision quantization is a promising approach for compressing large language models under tight memory budgets. However, existing mixed-precision methods typically suffer from one of two limitations: they either rely on expensive discrete optimization to determine precision allocation, or introduce hardware inefficiencies due to irregular memory layouts. We propose SFMP, a search-free and hardware-friendly mixed-precision quantization framework for large language models. The framework is built upon four novel ideas: Fractional bit-width, which extends integer bit-width for weight matrix to fractional value and transforms discrete precision allocation as a continuous problem; 2)Block-wise mixed-precision, enabling fine-grained precision within weight matrices while remaining hardware-friendly; 3)Row-column weight reordering, which aggregates salient weights via row and column reordering, incurring only a small activation reordering overhead during inference; 4)Unified GEMM kernel, which supports mixed-precision GEMM at arbitrary average bit-width. Extensive experiments demonstrate that SFMP outperforms state-of-the-art layer-wise mixed-precision methods under the same memory constraints, while significantly reducing quantization cost and improving inference efficiency. Code is available at https://github.com/Nkniexin/SFMP
HY-Motion 1.0: Scaling Flow Matching Models for Text-To-Motion Generation
Dec 29, 2025Abstract:We present HY-Motion 1.0, a series of state-of-the-art, large-scale, motion generation models capable of generating 3D human motions from textual descriptions. HY-Motion 1.0 represents the first successful attempt to scale up Diffusion Transformer (DiT)-based flow matching models to the billion-parameter scale within the motion generation domain, delivering instruction-following capabilities that significantly outperform current open-source benchmarks. Uniquely, we introduce a comprehensive, full-stage training paradigm -- including large-scale pretraining on over 3,000 hours of motion data, high-quality fine-tuning on 400 hours of curated data, and reinforcement learning from both human feedback and reward models -- to ensure precise alignment with the text instruction and high motion quality. This framework is supported by our meticulous data processing pipeline, which performs rigorous motion cleaning and captioning. Consequently, our model achieves the most extensive coverage, spanning over 200 motion categories across 6 major classes. We release HY-Motion 1.0 to the open-source community to foster future research and accelerate the transition of 3D human motion generation models towards commercial maturity.
Hunyuan3D Studio: End-to-End AI Pipeline for Game-Ready 3D Asset Generation
Sep 16, 2025



Abstract:The creation of high-quality 3D assets, a cornerstone of modern game development, has long been characterized by labor-intensive and specialized workflows. This paper presents Hunyuan3D Studio, an end-to-end AI-powered content creation platform designed to revolutionize the game production pipeline by automating and streamlining the generation of game-ready 3D assets. At its core, Hunyuan3D Studio integrates a suite of advanced neural modules (such as Part-level 3D Generation, Polygon Generation, Semantic UV, etc.) into a cohesive and user-friendly system. This unified framework allows for the rapid transformation of a single concept image or textual description into a fully-realized, production-quality 3D model complete with optimized geometry and high-fidelity PBR textures. We demonstrate that assets generated by Hunyuan3D Studio are not only visually compelling but also adhere to the stringent technical requirements of contemporary game engines, significantly reducing iteration time and lowering the barrier to entry for 3D content creation. By providing a seamless bridge from creative intent to technical asset, Hunyuan3D Studio represents a significant leap forward for AI-assisted workflows in game development and interactive media.
HunyuanWorld 1.0: Generating Immersive, Explorable, and Interactive 3D Worlds from Words or Pixels
Jul 29, 2025



Abstract:Creating immersive and playable 3D worlds from texts or images remains a fundamental challenge in computer vision and graphics. Existing world generation approaches typically fall into two categories: video-based methods that offer rich diversity but lack 3D consistency and rendering efficiency, and 3D-based methods that provide geometric consistency but struggle with limited training data and memory-inefficient representations. To address these limitations, we present HunyuanWorld 1.0, a novel framework that combines the best of both worlds for generating immersive, explorable, and interactive 3D scenes from text and image conditions. Our approach features three key advantages: 1) 360{\deg} immersive experiences via panoramic world proxies; 2) mesh export capabilities for seamless compatibility with existing computer graphics pipelines; 3) disentangled object representations for augmented interactivity. The core of our framework is a semantically layered 3D mesh representation that leverages panoramic images as 360{\deg} world proxies for semantic-aware world decomposition and reconstruction, enabling the generation of diverse 3D worlds. Extensive experiments demonstrate that our method achieves state-of-the-art performance in generating coherent, explorable, and interactive 3D worlds while enabling versatile applications in virtual reality, physical simulation, game development, and interactive content creation.
Enhancing Vehicular Platooning with Wireless Federated Learning: A Resource-Aware Control Framework
Jul 01, 2025Abstract:This paper aims to enhance the performance of Vehicular Platooning (VP) systems integrated with Wireless Federated Learning (WFL). In highly dynamic environments, vehicular platoons experience frequent communication changes and resource constraints, which significantly affect information exchange and learning model synchronization. To address these challenges, we first formulate WFL in VP as a joint optimization problem that simultaneously considers Age of Information (AoI) and Federated Learning Model Drift (FLMD) to ensure timely and accurate control. Through theoretical analysis, we examine the impact of FLMD on convergence performance and develop a two-stage Resource-Aware Control framework (RACE). The first stage employs a Lagrangian dual decomposition method for resource configuration, while the second stage implements a multi-agent deep reinforcement learning approach for vehicle selection. The approach integrates Multi-Head Self-Attention and Long Short-Term Memory networks to capture spatiotemporal correlations in communication states. Experimental results demonstrate that, compared to baseline methods, the proposed framework improves AoI optimization by up to 45%, accelerates learning convergence, and adapts more effectively to dynamic VP environments on the AI4MARS dataset.
Kling-Foley: Multimodal Diffusion Transformer for High-Quality Video-to-Audio Generation
Jun 24, 2025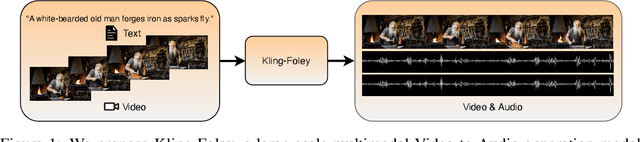
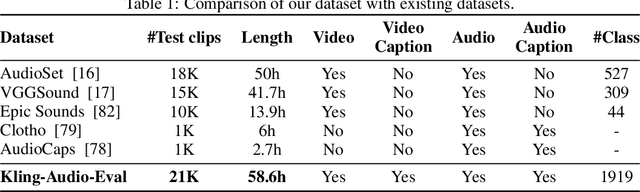
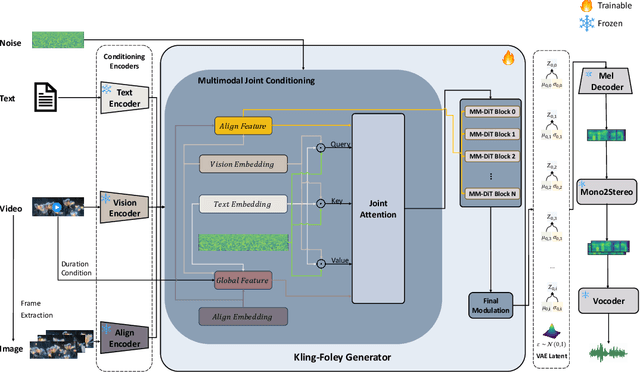
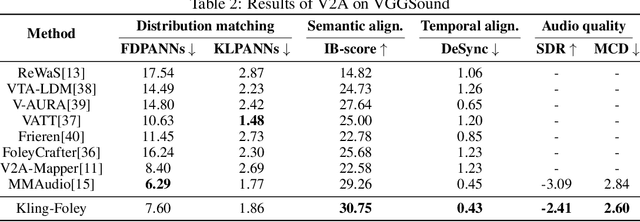
Abstract:We propose Kling-Foley, a large-scale multimodal Video-to-Audio generation model that synthesizes high-quality audio synchronized with video content. In Kling-Foley, we introduce multimodal diffusion transformers to model the interactions between video, audio, and text modalities, and combine it with a visual semantic representation module and an audio-visual synchronization module to enhance alignment capabilities. Specifically, these modules align video conditions with latent audio elements at the frame level, thereby improving semantic alignment and audio-visual synchronization. Together with text conditions, this integrated approach enables precise generation of video-matching sound effects. In addition, we propose a universal latent audio codec that can achieve high-quality modeling in various scenarios such as sound effects, speech, singing, and music. We employ a stereo rendering method that imbues synthesized audio with a spatial presence. At the same time, in order to make up for the incomplete types and annotations of the open-source benchmark, we also open-source an industrial-level benchmark Kling-Audio-Eval. Our experiments show that Kling-Foley trained with the flow matching objective achieves new audio-visual SOTA performance among public models in terms of distribution matching, semantic alignment, temporal alignment and audio quality.
Hunyuan3D 2.1: From Images to High-Fidelity 3D Assets with Production-Ready PBR Material
Jun 18, 2025Abstract:3D AI-generated content (AIGC) is a passionate field that has significantly accelerated the creation of 3D models in gaming, film, and design. Despite the development of several groundbreaking models that have revolutionized 3D generation, the field remains largely accessible only to researchers, developers, and designers due to the complexities involved in collecting, processing, and training 3D models. To address these challenges, we introduce Hunyuan3D 2.1 as a case study in this tutorial. This tutorial offers a comprehensive, step-by-step guide on processing 3D data, training a 3D generative model, and evaluating its performance using Hunyuan3D 2.1, an advanced system for producing high-resolution, textured 3D assets. The system comprises two core components: the Hunyuan3D-DiT for shape generation and the Hunyuan3D-Paint for texture synthesis. We will explore the entire workflow, including data preparation, model architecture, training strategies, evaluation metrics, and deployment. By the conclusion of this tutorial, you will have the knowledge to finetune or develop a robust 3D generative model suitable for applications in gaming, virtual reality, and industrial design.
Hunyuan-TurboS: Advancing Large Language Models through Mamba-Transformer Synergy and Adaptive Chain-of-Thought
May 21, 2025Abstract:As Large Language Models (LLMs) rapidly advance, we introduce Hunyuan-TurboS, a novel large hybrid Transformer-Mamba Mixture of Experts (MoE) model. It synergistically combines Mamba's long-sequence processing efficiency with Transformer's superior contextual understanding. Hunyuan-TurboS features an adaptive long-short chain-of-thought (CoT) mechanism, dynamically switching between rapid responses for simple queries and deep "thinking" modes for complex problems, optimizing computational resources. Architecturally, this 56B activated (560B total) parameter model employs 128 layers (Mamba2, Attention, FFN) with an innovative AMF/MF block pattern. Faster Mamba2 ensures linear complexity, Grouped-Query Attention minimizes KV cache, and FFNs use an MoE structure. Pre-trained on 16T high-quality tokens, it supports a 256K context length and is the first industry-deployed large-scale Mamba model. Our comprehensive post-training strategy enhances capabilities via Supervised Fine-Tuning (3M instructions), a novel Adaptive Long-short CoT Fusion method, Multi-round Deliberation Learning for iterative improvement, and a two-stage Large-scale Reinforcement Learning process targeting STEM and general instruction-following. Evaluations show strong performance: overall top 7 rank on LMSYS Chatbot Arena with a score of 1356, outperforming leading models like Gemini-2.0-Flash-001 (1352) and o4-mini-2025-04-16 (1345). TurboS also achieves an average of 77.9% across 23 automated benchmarks. Hunyuan-TurboS balances high performance and efficiency, offering substantial capabilities at lower inference costs than many reasoning models, establishing a new paradigm for efficient large-scale pre-trained models.
Neural Pruning for 3D Scene Reconstruction: Efficient NeRF Acceleration
Apr 01, 2025Abstract:Neural Radiance Fields (NeRF) have become a popular 3D reconstruction approach in recent years. While they produce high-quality results, they also demand lengthy training times, often spanning days. This paper studies neural pruning as a strategy to address these concerns. We compare pruning approaches, including uniform sampling, importance-based methods, and coreset-based techniques, to reduce the model size and speed up training. Our findings show that coreset-driven pruning can achieve a 50% reduction in model size and a 35% speedup in training, with only a slight decrease in accuracy. These results suggest that pruning can be an effective method for improving the efficiency of NeRF models in resource-limited settings.
GKAN: Explainable Diagnosis of Alzheimer's Disease Using Graph Neural Network with Kolmogorov-Arnold Networks
Apr 01, 2025Abstract:Alzheimer's Disease (AD) is a progressive neurodegenerative disorder that poses significant diagnostic challenges due to its complex etiology. Graph Convolutional Networks (GCNs) have shown promise in modeling brain connectivity for AD diagnosis, yet their reliance on linear transformations limits their ability to capture intricate nonlinear patterns in neuroimaging data. To address this, we propose GCN-KAN, a novel single-modal framework that integrates Kolmogorov-Arnold Networks (KAN) into GCNs to enhance both diagnostic accuracy and interpretability. Leveraging structural MRI data, our model employs learnable spline-based transformations to better represent brain region interactions. Evaluated on the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, GCN-KAN outperforms traditional GCNs by 4-8% in classification accuracy while providing interpretable insights into key brain regions associated with AD. This approach offers a robust and explainable tool for early AD diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge