Lei Li
Carnegie Mellon University
On uniform-in-time diffusion approximation for stochastic gradient descent
Jul 11, 2022Abstract:The diffusion approximation of stochastic gradient descent (SGD) in current literature is only valid on a finite time interval. In this paper, we establish the uniform-in-time diffusion approximation of SGD, by only assuming that the expected loss is strongly convex and some other mild conditions, without assuming the convexity of each random loss function. The main technique is to establish the exponential decay rates of the derivatives of the solution to the backward Kolmogorov equation. The uniform-in-time approximation allows us to study asymptotic behaviors of SGD via the continuous stochastic differential equation (SDE) even when the random objective function $f(\cdot;\xi)$ is not strongly convex.
On the Impact of Noises in Crowd-Sourced Data for Speech Translation
Jul 01, 2022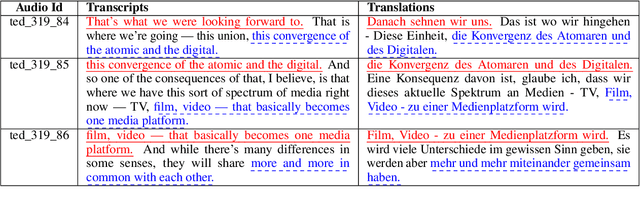

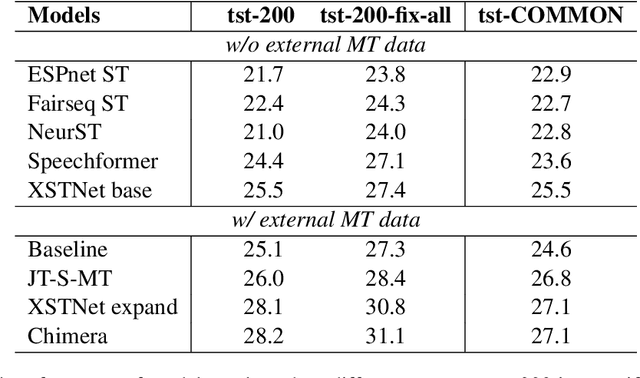
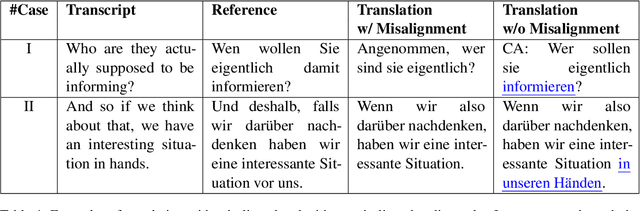
Abstract:Training speech translation (ST) models requires large and high-quality datasets. MuST-C is one of the most widely used ST benchmark datasets. It contains around 400 hours of speech-transcript-translation data for each of the eight translation directions. This dataset passes several quality-control filters during creation. However, we find that MuST-C still suffers from three major quality issues: audio-text misalignment, inaccurate translation, and unnecessary speaker's name. What are the impacts of these data quality issues for model development and evaluation? In this paper, we propose an automatic method to fix or filter the above quality issues, using English-German (En-De) translation as an example. Our experiments show that ST models perform better on clean test sets, and the rank of proposed models remains consistent across different test sets. Besides, simply removing misaligned data points from the training set does not lead to a better ST model.
CSI Sensing from Heterogeneous User Feedbacks: A Constrained Phase Retrieval Approach
Jun 28, 2022
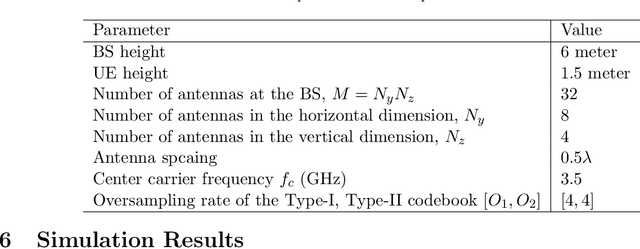
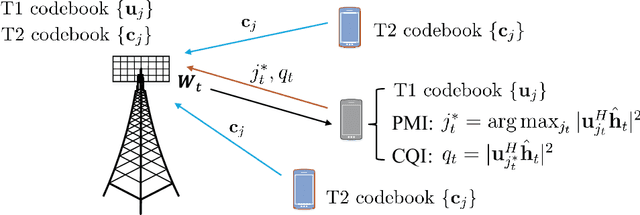
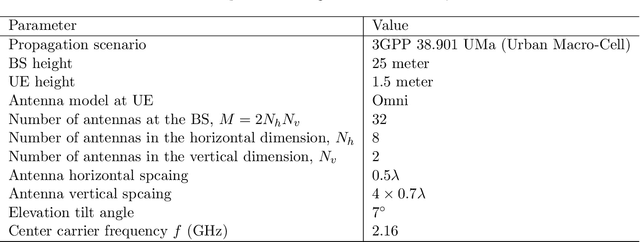
Abstract:This paper investigates the downlink channel state information (CSI) sensing in 5G heterogeneous networks composed of user equipments (UEs) with different feedback capabilities. We aim to enhance the CSI accuracy of UEs only affording the low-resolution Type-I codebook. While existing works have demonstrated that the task can be accomplished by solving a phase retrieval (PR) formulation based on the feedback of precoding matrix indicator (PMI) and channel quality indicator (CQI), they need many feedback rounds. In this paper, we propose a novel CSI sensing scheme that can significantly reduce the feedback overhead. Our scheme involves a novel parameter dimension reduction design by exploiting the spatial consistency of wireless channels among nearby UEs, and a constrained PR (CPR) formulation that characterizes the feasible region of CSI by the PMI information. To address the computational challenge due to the non-convexity and the large number of constraints of CPR, we develop a two-stage algorithm that firstly identifies and removes inactive constraints, followed by a fast first-order algorithm. The study is further extended to multi-carrier systems. Extensive tests over DeepMIMO and QuaDriGa datasets showcase that our designs greatly outperform existing methods and achieve the high-resolution Type-II codebook performance with a few rounds of feedback.
On the Learning of Non-Autoregressive Transformers
Jun 13, 2022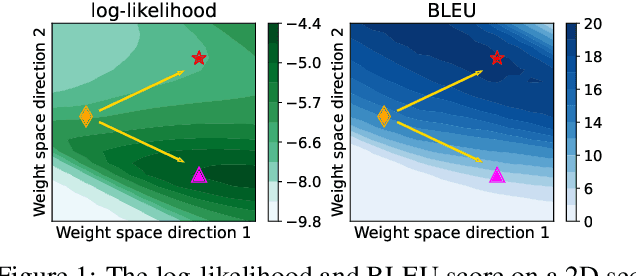
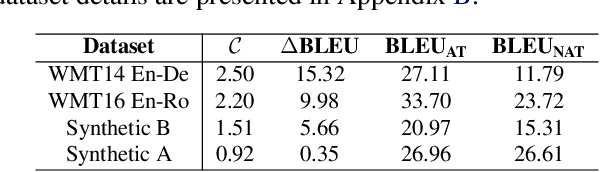
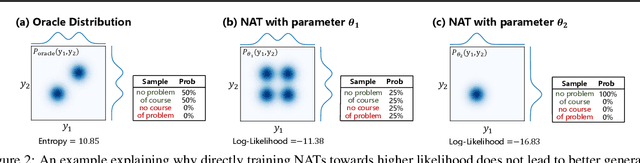
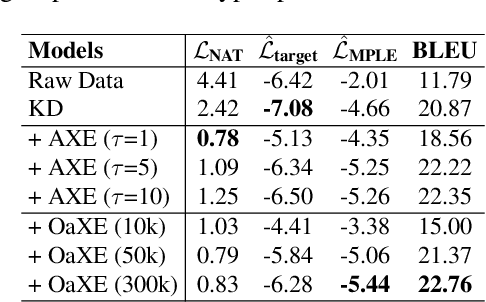
Abstract:Non-autoregressive Transformer (NAT) is a family of text generation models, which aims to reduce the decoding latency by predicting the whole sentences in parallel. However, such latency reduction sacrifices the ability to capture left-to-right dependencies, thereby making NAT learning very challenging. In this paper, we present theoretical and empirical analyses to reveal the challenges of NAT learning and propose a unified perspective to understand existing successes. First, we show that simply training NAT by maximizing the likelihood can lead to an approximation of marginal distributions but drops all dependencies between tokens, where the dropped information can be measured by the dataset's conditional total correlation. Second, we formalize many previous objectives in a unified framework and show that their success can be concluded as maximizing the likelihood on a proxy distribution, leading to a reduced information loss. Empirical studies show that our perspective can explain the phenomena in NAT learning and guide the design of new training methods.
Decoupling Predictions in Distributed Learning for Multi-Center Left Atrial MRI Segmentation
Jun 10, 2022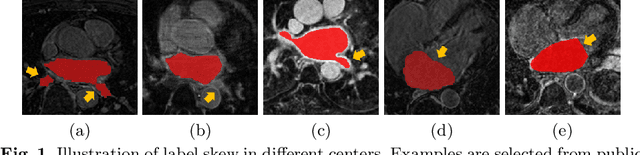
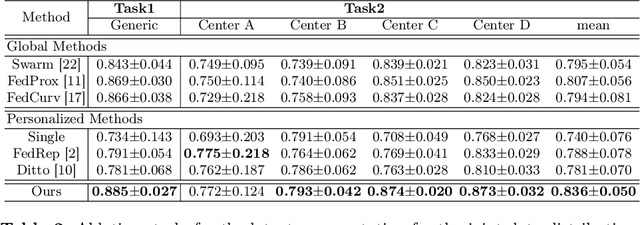
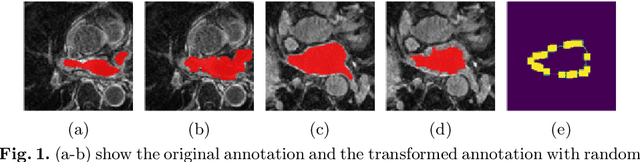
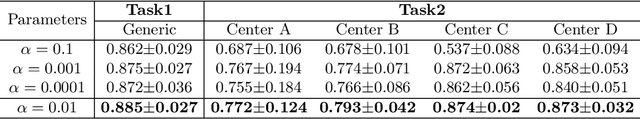
Abstract:Distributed learning has shown great potential in medical image analysis. It allows to use multi-center training data with privacy protection. However, data distributions in local centers can vary from each other due to different imaging vendors, and annotation protocols. Such variation degrades the performance of learning-based methods. To mitigate the influence, two groups of methods have been proposed for different aims, i.e., the global methods and the personalized methods. The former are aimed to improve the performance of a single global model for all test data from unseen centers (known as generic data); while the latter target multiple models for each center (denoted as local data). However, little has been researched to achieve both goals simultaneously. In this work, we propose a new framework of distributed learning that bridges the gap between two groups, and improves the performance for both generic and local data. Specifically, our method decouples the predictions for generic data and local data, via distribution-conditioned adaptation matrices. Results on multi-center left atrial (LA) MRI segmentation showed that our method demonstrated superior performance over existing methods on both generic and local data. Our code is available at https://github.com/key1589745/decouple_predict
Rethinking the Openness of CLIP
Jun 04, 2022Abstract:Contrastive Language-Image Pre-training (CLIP) has demonstrated great potential in realizing open-vocabulary image classification in a matching style, because of its holistic use of natural language supervision that covers unconstrained real-world visual concepts. However, it is, in turn, also difficult to evaluate and analyze the openness of CLIP-like models, since they are in theory open to any vocabulary but the actual accuracy varies. To address the insufficiency of conventional studies on openness, we resort to an incremental view and define the extensibility, which essentially approximates the model's ability to deal with new visual concepts, by evaluating openness through vocabulary expansions. Our evaluation based on extensibility shows that CLIP-like models are hardly truly open and their performances degrade as the vocabulary expands to different degrees. Further analysis reveals that the over-estimation of openness is not because CLIP-like models fail to capture the general similarity of image and text features of novel visual concepts, but because of the confusion among competing text features, that is, they are not stable with respect to the vocabulary. In light of this, we propose to improve the openness of CLIP from the perspective of feature space by enforcing the distinguishability of text features. Our method retrieves relevant texts from the pre-training corpus to enhance prompts for inference, which boosts the extensibility and stability of CLIP even without fine-tuning.
Decoupling Knowledge from Memorization: Retrieval-augmented Prompt Learning
May 29, 2022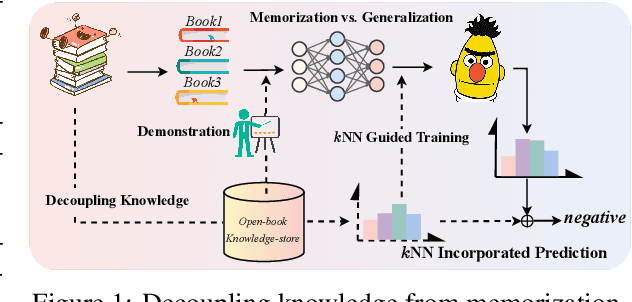
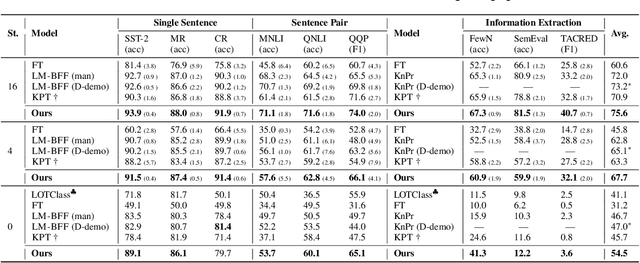
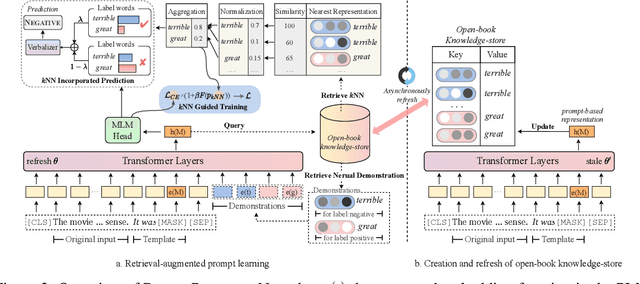
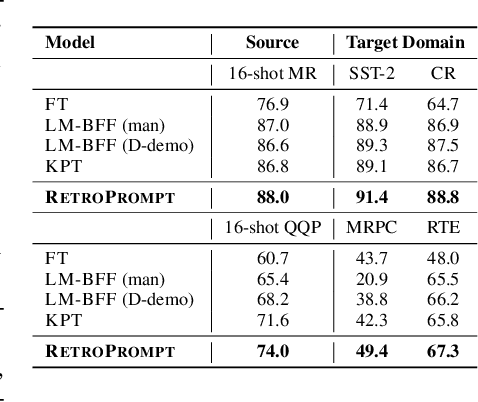
Abstract:Prompt learning approaches have made waves in natural language processing by inducing better few-shot performance while they still follow a parametric-based learning paradigm; the oblivion and rote memorization problems in learning may encounter unstable generalization issues. Specifically, vanilla prompt learning may struggle to utilize atypical instances by rote during fully-supervised training or overfit shallow patterns with low-shot data. To alleviate such limitations, we develop RetroPrompt with the motivation of decoupling knowledge from memorization to help the model strike a balance between generalization and memorization. In contrast with vanilla prompt learning, RetroPrompt constructs an open-book knowledge-store from training instances and implements a retrieval mechanism during the process of input, training and inference, thus equipping the model with the ability to retrieve related contexts from the training corpus as cues for enhancement. Extensive experiments demonstrate that RetroPrompt can obtain better performance in both few-shot and zero-shot settings. Besides, we further illustrate that our proposed RetroPrompt can yield better generalization abilities with new datasets. Detailed analysis of memorization indeed reveals RetroPrompt can reduce the reliance of language models on memorization; thus, improving generalization for downstream tasks.
Enhancing Cross-lingual Transfer by Manifold Mixup
May 09, 2022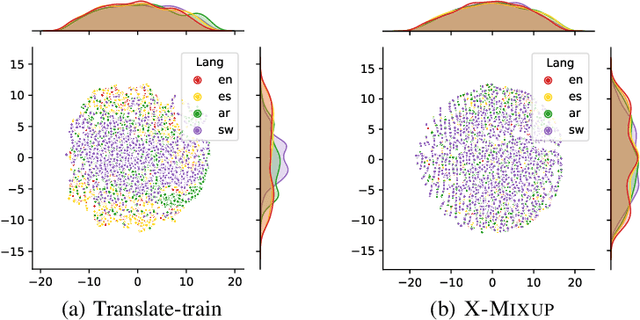
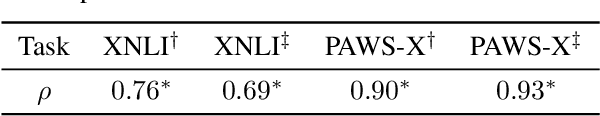
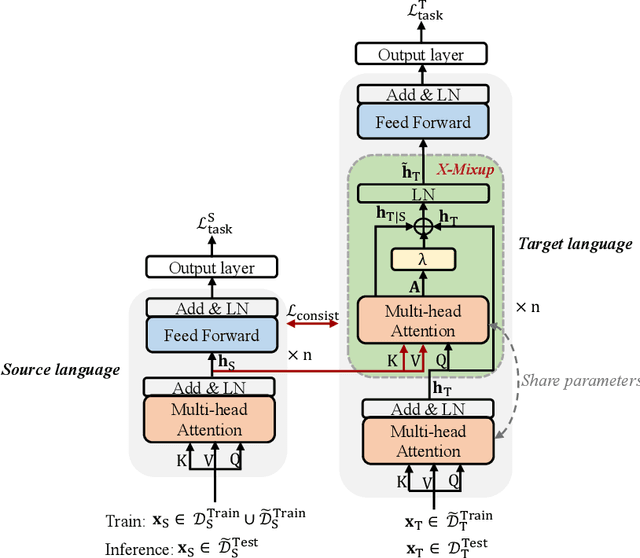
Abstract:Based on large-scale pre-trained multilingual representations, recent cross-lingual transfer methods have achieved impressive transfer performances. However, the performance of target languages still lags far behind the source language. In this paper, our analyses indicate such a performance gap is strongly associated with the cross-lingual representation discrepancy. To achieve better cross-lingual transfer performance, we propose the cross-lingual manifold mixup (X-Mixup) method, which adaptively calibrates the representation discrepancy and gives a compromised representation for target languages. Experiments on the XTREME benchmark show X-Mixup achieves 1.8% performance gains on multiple text understanding tasks, compared with strong baselines, and significantly reduces the cross-lingual representation discrepancy.
Good Visual Guidance Makes A Better Extractor: Hierarchical Visual Prefix for Multimodal Entity and Relation Extraction
May 07, 2022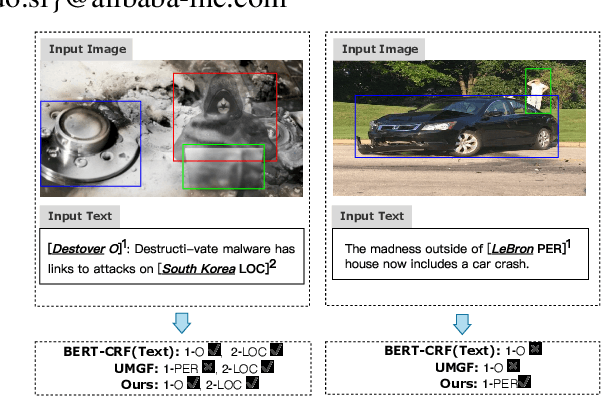
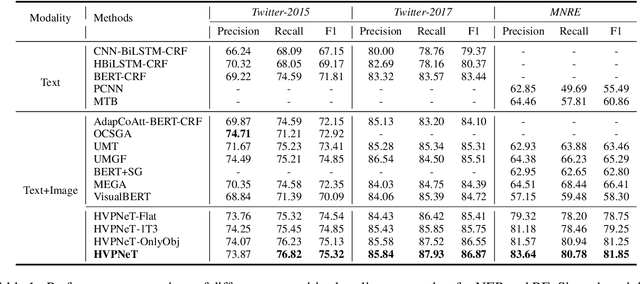
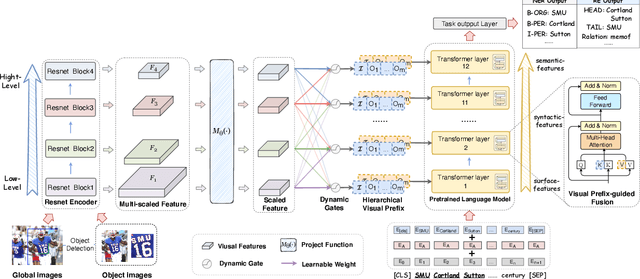
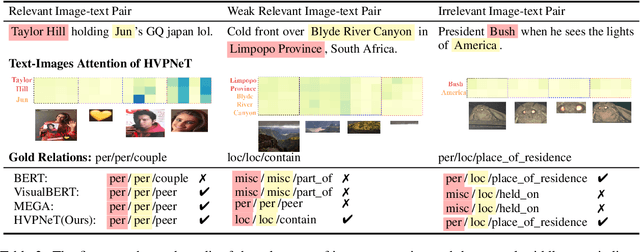
Abstract:Multimodal named entity recognition and relation extraction (MNER and MRE) is a fundamental and crucial branch in information extraction. However, existing approaches for MNER and MRE usually suffer from error sensitivity when irrelevant object images incorporated in texts. To deal with these issues, we propose a novel Hierarchical Visual Prefix fusion NeTwork (HVPNeT) for visual-enhanced entity and relation extraction, aiming to achieve more effective and robust performance. Specifically, we regard visual representation as pluggable visual prefix to guide the textual representation for error insensitive forecasting decision. We further propose a dynamic gated aggregation strategy to achieve hierarchical multi-scaled visual features as visual prefix for fusion. Extensive experiments on three benchmark datasets demonstrate the effectiveness of our method, and achieve state-of-the-art performance. Code is available in https://github.com/zjunlp/HVPNeT.
Cross-modal Contrastive Learning for Speech Translation
May 05, 2022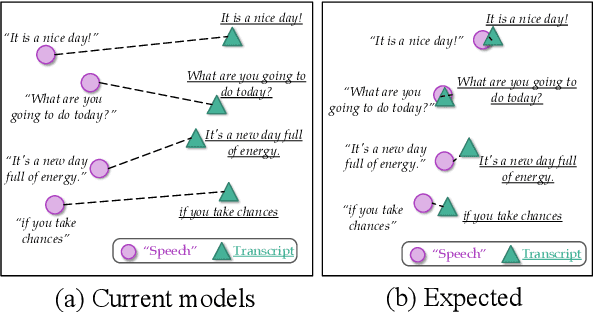
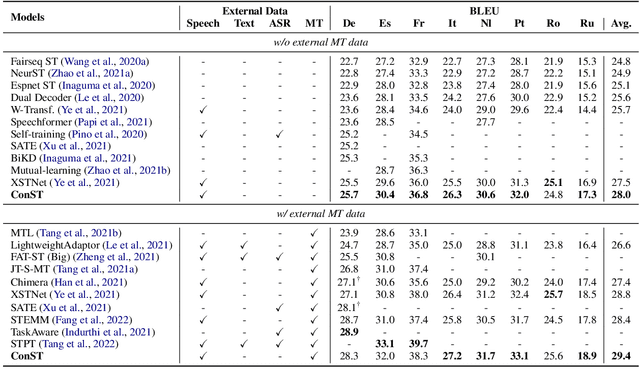
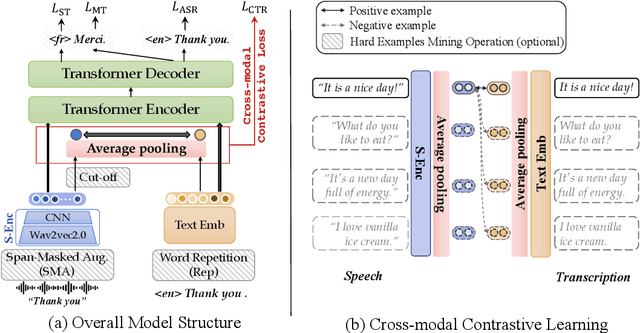
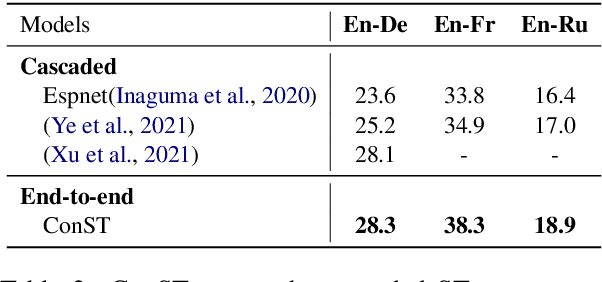
Abstract:How can we learn unified representations for spoken utterances and their written text? Learning similar representations for semantically similar speech and text is important for speech translation. To this end, we propose ConST, a cross-modal contrastive learning method for end-to-end speech-to-text translation. We evaluate ConST and a variety of previous baselines on a popular benchmark MuST-C. Experiments show that the proposed ConST consistently outperforms the previous methods on, and achieves an average BLEU of 29.4. The analysis further verifies that ConST indeed closes the representation gap of different modalities -- its learned representation improves the accuracy of cross-modal speech-text retrieval from 4% to 88%. Code and models are available at https://github.com/ReneeYe/ConST.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge