Yihao Chen
ScaleEnv: Scaling Environment Synthesis from Scratch for Generalist Interactive Tool-Use Agent Training
Feb 06, 2026Abstract:Training generalist agents capable of adapting to diverse scenarios requires interactive environments for self-exploration. However, interactive environments remain critically scarce, and existing synthesis methods suffer from significant limitations regarding environmental diversity and scalability. To address these challenges, we introduce ScaleEnv, a framework that constructs fully interactive environments and verifiable tasks entirely from scratch. Specifically, ScaleEnv ensures environment reliability through procedural testing, and guarantees task completeness and solvability via tool dependency graph expansion and executable action verification. By enabling agents to learn through exploration within ScaleEnv, we demonstrate significant performance improvements on unseen, multi-turn tool-use benchmarks such as $τ^2$-Bench and VitaBench, highlighting strong generalization capabilities. Furthermore, we investigate the relationship between increasing number of domains and model generalization performance, providing empirical evidence that scaling environmental diversity is critical for robust agent learning.
LongCat-Flash-Thinking-2601 Technical Report
Jan 23, 2026Abstract:We introduce LongCat-Flash-Thinking-2601, a 560-billion-parameter open-source Mixture-of-Experts (MoE) reasoning model with superior agentic reasoning capability. LongCat-Flash-Thinking-2601 achieves state-of-the-art performance among open-source models on a wide range of agentic benchmarks, including agentic search, agentic tool use, and tool-integrated reasoning. Beyond benchmark performance, the model demonstrates strong generalization to complex tool interactions and robust behavior under noisy real-world environments. Its advanced capability stems from a unified training framework that combines domain-parallel expert training with subsequent fusion, together with an end-to-end co-design of data construction, environments, algorithms, and infrastructure spanning from pre-training to post-training. In particular, the model's strong generalization capability in complex tool-use are driven by our in-depth exploration of environment scaling and principled task construction. To optimize long-tailed, skewed generation and multi-turn agentic interactions, and to enable stable training across over 10,000 environments spanning more than 20 domains, we systematically extend our asynchronous reinforcement learning framework, DORA, for stable and efficient large-scale multi-environment training. Furthermore, recognizing that real-world tasks are inherently noisy, we conduct a systematic analysis and decomposition of real-world noise patterns, and design targeted training procedures to explicitly incorporate such imperfections into the training process, resulting in improved robustness for real-world applications. To further enhance performance on complex reasoning tasks, we introduce a Heavy Thinking mode that enables effective test-time scaling by jointly expanding reasoning depth and width through intensive parallel thinking.
Detect Anything via Next Point Prediction
Oct 14, 2025Abstract:Object detection has long been dominated by traditional coordinate regression-based models, such as YOLO, DETR, and Grounding DINO. Although recent efforts have attempted to leverage MLLMs to tackle this task, they face challenges like low recall rate, duplicate predictions, coordinate misalignment, etc. In this work, we bridge this gap and propose Rex-Omni, a 3B-scale MLLM that achieves state-of-the-art object perception performance. On benchmarks like COCO and LVIS, Rex-Omni attains performance comparable to or exceeding regression-based models (e.g., DINO, Grounding DINO) in a zero-shot setting. This is enabled by three key designs: 1) Task Formulation: we use special tokens to represent quantized coordinates from 0 to 999, reducing the model's learning difficulty and improving token efficiency for coordinate prediction; 2) Data Engines: we construct multiple data engines to generate high-quality grounding, referring, and pointing data, providing semantically rich supervision for training; \3) Training Pipelines: we employ a two-stage training process, combining supervised fine-tuning on 22 million data with GRPO-based reinforcement post-training. This RL post-training leverages geometry-aware rewards to effectively bridge the discrete-to-continuous coordinate prediction gap, improve box accuracy, and mitigate undesirable behaviors like duplicate predictions that stem from the teacher-guided nature of the initial SFT stage. Beyond conventional detection, Rex-Omni's inherent language understanding enables versatile capabilities such as object referring, pointing, visual prompting, GUI grounding, spatial referring, OCR and key-pointing, all systematically evaluated on dedicated benchmarks. We believe that Rex-Omni paves the way for more versatile and language-aware visual perception systems.
Physics-Constrained Diffusion Reconstruction with Posterior Correction for Quantitative and Fast PET Imaging
Aug 20, 2025



Abstract:Deep learning-based reconstruction of positron emission tomography(PET) data has gained increasing attention in recent years. While these methods achieve fast reconstruction,concerns remain regarding quantitative accuracy and the presence of artifacts,stemming from limited model interpretability,data driven dependence, and overfitting risks.These challenges have hindered clinical adoption.To address them,we propose a conditional diffusion model with posterior physical correction (PET-DPC) for PET image reconstruction. An innovative normalization procedure generates the input Geometric TOF Probabilistic Image (GTP-image),while physical information is incorporated during the diffusion sampling process to perform posterior scatter,attenuation,and random corrections. The model was trained and validated on 300 brain and 50 whole-body PET datasets,a physical phantom,and 20 simulated brain datasets. PET-DPC produced reconstructions closely aligned with fully corrected OSEM images,outperforming end-to-end deep learning models in quantitative metrics and,in some cases, surpassing traditional iterative methods. The model also generalized well to out-of-distribution(OOD) data. Compared to iterative methods,PET-DPC reduced reconstruction time by 50% for brain scans and 85% for whole-body scans. Ablation studies confirmed the critical role of posterior correction in implementing scatter and attenuation corrections,enhancing reconstruction accuracy. Experiments with physical phantoms further demonstrated PET-DPC's ability to preserve background uniformity and accurately reproduce tumor-to-background intensity ratios. Overall,these results highlight PET-DPC as a promising approach for rapid, quantitatively accurate PET reconstruction,with strong potential to improve clinical imaging workflows.
The Fourth Monocular Depth Estimation Challenge
Apr 24, 2025
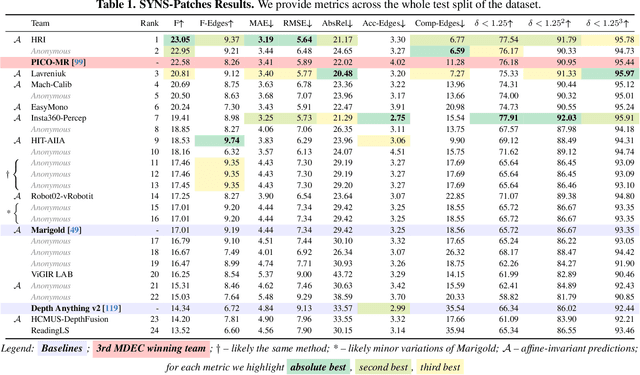
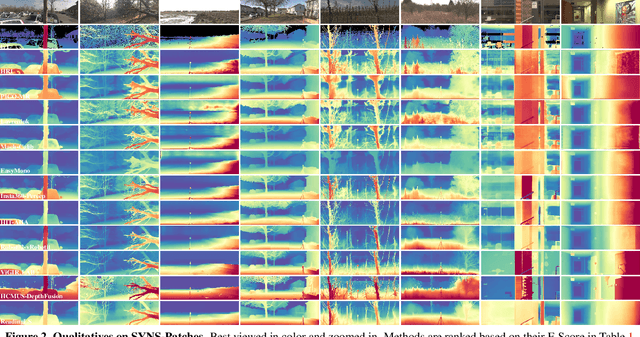
Abstract:This paper presents the results of the fourth edition of the Monocular Depth Estimation Challenge (MDEC), which focuses on zero-shot generalization to the SYNS-Patches benchmark, a dataset featuring challenging environments in both natural and indoor settings. In this edition, we revised the evaluation protocol to use least-squares alignment with two degrees of freedom to support disparity and affine-invariant predictions. We also revised the baselines and included popular off-the-shelf methods: Depth Anything v2 and Marigold. The challenge received a total of 24 submissions that outperformed the baselines on the test set; 10 of these included a report describing their approach, with most leading methods relying on affine-invariant predictions. The challenge winners improved the 3D F-Score over the previous edition's best result, raising it from 22.58% to 23.05%.
Referring to Any Person
Mar 11, 2025
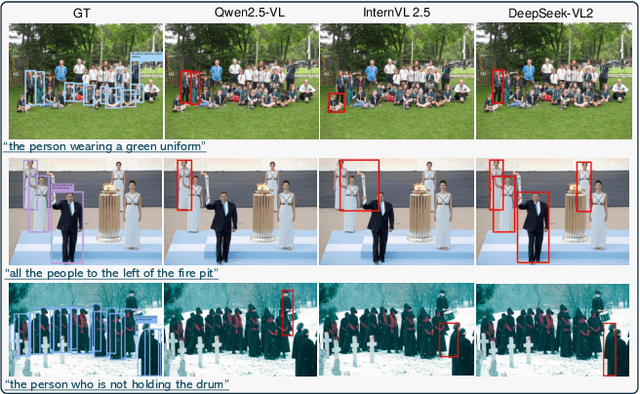
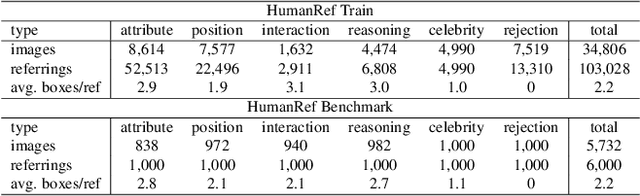
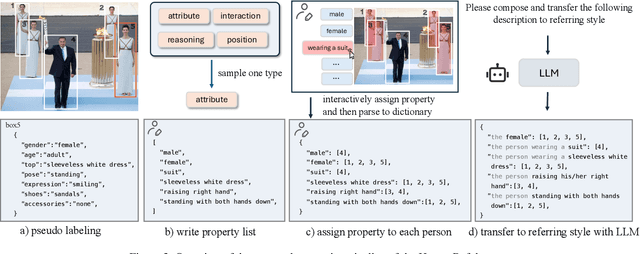
Abstract:Humans are undoubtedly the most important participants in computer vision, and the ability to detect any individual given a natural language description, a task we define as referring to any person, holds substantial practical value. However, we find that existing models generally fail to achieve real-world usability, and current benchmarks are limited by their focus on one-to-one referring, that hinder progress in this area. In this work, we revisit this task from three critical perspectives: task definition, dataset design, and model architecture. We first identify five aspects of referable entities and three distinctive characteristics of this task. Next, we introduce HumanRef, a novel dataset designed to tackle these challenges and better reflect real-world applications. From a model design perspective, we integrate a multimodal large language model with an object detection framework, constructing a robust referring model named RexSeek. Experimental results reveal that state-of-the-art models, which perform well on commonly used benchmarks like RefCOCO/+/g, struggle with HumanRef due to their inability to detect multiple individuals. In contrast, RexSeek not only excels in human referring but also generalizes effectively to common object referring, making it broadly applicable across various perception tasks. Code is available at https://github.com/IDEA-Research/RexSeek
ChatRex: Taming Multimodal LLM for Joint Perception and Understanding
Dec 02, 2024



Abstract:Perception and understanding are two pillars of computer vision. While multimodal large language models (MLLM) have demonstrated remarkable visual understanding capabilities, they arguably lack accurate perception abilities, e.g. the stage-of-the-art model Qwen2-VL only achieves a 43.9 recall rate on the COCO dataset, limiting many tasks requiring the combination of perception and understanding. In this work, we aim to bridge this perception gap from both model designing and data development perspectives. We first introduce ChatRex, an MLLM with a decoupled perception design. Instead of having the LLM directly predict box coordinates, we feed the output boxes from a universal proposal network into the LLM, allowing it to output the corresponding box indices to represent its detection results, turning the regression task into a retrieval-based task that LLM handles more proficiently. From the data perspective, we build a fully automated data engine and construct the Rexverse-2M dataset which possesses multiple granularities to support the joint training of perception and understanding. After standard two-stage training, ChatRex demonstrates strong perception capabilities while preserving multimodal understanding performance. The combination of these two capabilities simultaneously unlocks many attractive applications, demonstrating the complementary roles of both perception and understanding in MLLM. Code is available at \url{https://github.com/IDEA-Research/ChatRex}.
DINO-X: A Unified Vision Model for Open-World Object Detection and Understanding
Nov 21, 2024



Abstract:In this paper, we introduce DINO-X, which is a unified object-centric vision model developed by IDEA Research with the best open-world object detection performance to date. DINO-X employs the same Transformer-based encoder-decoder architecture as Grounding DINO 1.5 to pursue an object-level representation for open-world object understanding. To make long-tailed object detection easy, DINO-X extends its input options to support text prompt, visual prompt, and customized prompt. With such flexible prompt options, we develop a universal object prompt to support prompt-free open-world detection, making it possible to detect anything in an image without requiring users to provide any prompt. To enhance the model's core grounding capability, we have constructed a large-scale dataset with over 100 million high-quality grounding samples, referred to as Grounding-100M, for advancing the model's open-vocabulary detection performance. Pre-training on such a large-scale grounding dataset leads to a foundational object-level representation, which enables DINO-X to integrate multiple perception heads to simultaneously support multiple object perception and understanding tasks, including detection, segmentation, pose estimation, object captioning, object-based QA, etc. Experimental results demonstrate the superior performance of DINO-X. Specifically, the DINO-X Pro model achieves 56.0 AP, 59.8 AP, and 52.4 AP on the COCO, LVIS-minival, and LVIS-val zero-shot object detection benchmarks, respectively. Notably, it scores 63.3 AP and 56.5 AP on the rare classes of LVIS-minival and LVIS-val benchmarks, both improving the previous SOTA performance by 5.8 AP. Such a result underscores its significantly improved capacity for recognizing long-tailed objects.
SGLP: A Similarity Guided Fast Layer Partition Pruning for Compressing Large Deep Models
Oct 14, 2024



Abstract:The deployment of Deep Neural Network (DNN)-based networks on resource-constrained devices remains a significant challenge due to their high computational and parameter requirements. To solve this problem, layer pruning has emerged as a potent approach to reduce network size and improve computational efficiency. However, existing layer pruning methods mostly overlook the intrinsic connections and inter-dependencies between different layers within complicated deep neural networks. This oversight can result in pruned models that do not preserve the essential characteristics of the pre-trained network as effectively as desired. To address this limitations, we propose a Similarity Guided fast Layer Partition pruning for compressing large deep models (SGLP), which focuses on pruning layers from network segments partitioned via representation similarity. Specifically, our presented method first leverages Centered Kernel Alignment (CKA) to indicate the internal representations among the layers of the pre-trained network, which provides us with a potent basis for layer pruning. Based on similarity matrix derived from CKA, we employ Fisher Optimal Segmentation to partition the network into multiple segments, which provides a basis for removing the layers in a segment-wise manner. In addition, our method innovatively adopts GradNorm for segment-wise layer importance evaluation, eliminating the need for extensive fine-tuning, and finally prunes the unimportant layers to obtain a compact network. Experimental results in image classification and for large language models (LLMs) demonstrate that our proposed SGLP outperforms the state-of-the-art methods in both accuracy and computational efficiency, presenting a more effective solution for deploying DNNs on resource-limited platforms. Our codes are available at https://github.com/itsnotacie/information-fusion-SGLP.
Deep reinforcement learning for tracking a moving target in jellyfish-like swimming
Sep 13, 2024



Abstract:We develop a deep reinforcement learning method for training a jellyfish-like swimmer to effectively track a moving target in a two-dimensional flow. This swimmer is a flexible object equipped with a muscle model based on torsional springs. We employ a deep Q-network (DQN) that takes the swimmer's geometry and dynamic parameters as inputs, and outputs actions which are the forces applied to the swimmer. In particular, we introduce an action regulation to mitigate the interference from complex fluid-structure interactions. The goal of these actions is to navigate the swimmer to a target point in the shortest possible time. In the DQN training, the data on the swimmer's motions are obtained from simulations conducted using the immersed boundary method. During tracking a moving target, there is an inherent delay between the application of forces and the corresponding response of the swimmer's body due to hydrodynamic interactions between the shedding vortices and the swimmer's own locomotion. Our tests demonstrate that the swimmer, with the DQN agent and action regulation, is able to dynamically adjust its course based on its instantaneous state. This work extends the application scope of machine learning in controlling flexible objects within fluid environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge