Huiyu Zhou
Cross-Skeleton Interaction Graph Aggregation Network for Representation Learning of Mouse Social Behaviour
Aug 07, 2022



Abstract:Automated social behaviour analysis of mice has become an increasingly popular research area in behavioural neuroscience. Recently, pose information (i.e., locations of keypoints or skeleton) has been used to interpret social behaviours of mice. Nevertheless, effective encoding and decoding of social interaction information underlying the keypoints of mice has been rarely investigated in the existing methods. In particular, it is challenging to model complex social interactions between mice due to highly deformable body shapes and ambiguous movement patterns. To deal with the interaction modelling problem, we here propose a Cross-Skeleton Interaction Graph Aggregation Network (CS-IGANet) to learn abundant dynamics of freely interacting mice, where a Cross-Skeleton Node-level Interaction module (CS-NLI) is used to model multi-level interactions (i.e., intra-, inter- and cross-skeleton interactions). Furthermore, we design a novel Interaction-Aware Transformer (IAT) to dynamically learn the graph-level representation of social behaviours and update the node-level representation, guided by our proposed interaction-aware self-attention mechanism. Finally, to enhance the representation ability of our model, an auxiliary self-supervised learning task is proposed for measuring the similarity between cross-skeleton nodes. Experimental results on the standard CRMI13-Skeleton and our PDMB-Skeleton datasets show that our proposed model outperforms several other state-of-the-art approaches.
giMLPs: Gate with Inhibition Mechanism in MLPs
Aug 02, 2022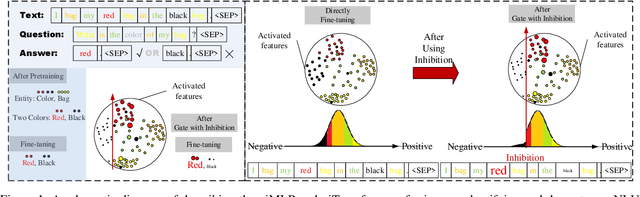
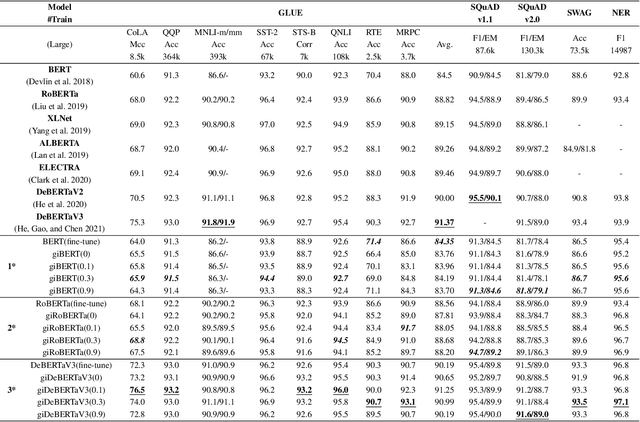
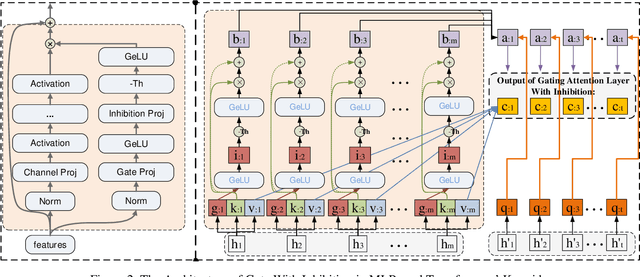
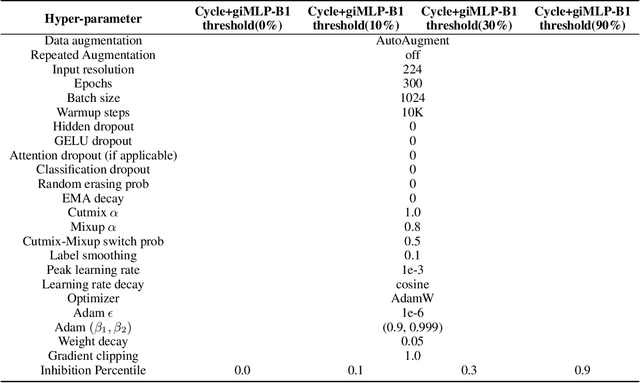
Abstract:This paper presents a new model architecture, gate with inhibition MLP (giMLP).The gate with inhibition on CycleMLP (gi-CycleMLP) can produce equal performance on the ImageNet classification task, and it also improves the BERT, Roberta, and DeBERTaV3 models depending on two novel techniques. The first is the gating MLP, where matrix multiplications between the MLP and the trunk Attention input in further adjust models' adaptation. The second is inhibition which inhibits or enhances the branch adjustment, and with the inhibition levels increasing, it offers models more muscular features restriction. We show that the giCycleMLP with a lower inhibition level can be competitive with the original CycleMLP in terms of ImageNet classification accuracy. In addition, we also show through a comprehensive empirical study that these techniques significantly improve the performance of fine-tuning NLU downstream tasks. As for the gate with inhibition MLPs on DeBERTa (giDeBERTa) fine-tuning, we find it can achieve appealing results on most parts of NLU tasks without any extra pretraining again. We also find that with the use of Gate With Inhibition, the activation function should have a short and smooth negative tail, with which the unimportant features or the features that hurt models can be moderately inhibited. The experiments on ImageNet and twelve language downstream tasks demonstrate the effectiveness of Gate With Inhibition, both for image classification and for enhancing the capacity of nature language fine-tuning without any extra pretraining.
CTooth+: A Large-scale Dental Cone Beam Computed Tomography Dataset and Benchmark for Tooth Volume Segmentation
Aug 02, 2022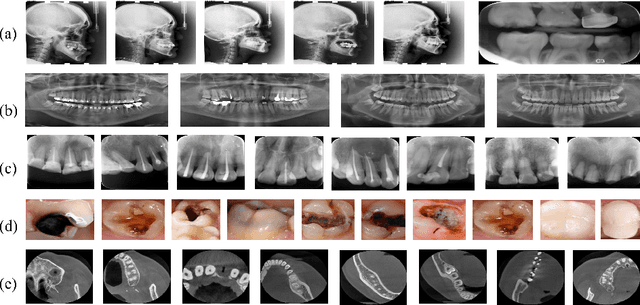
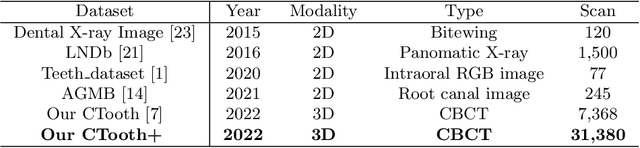
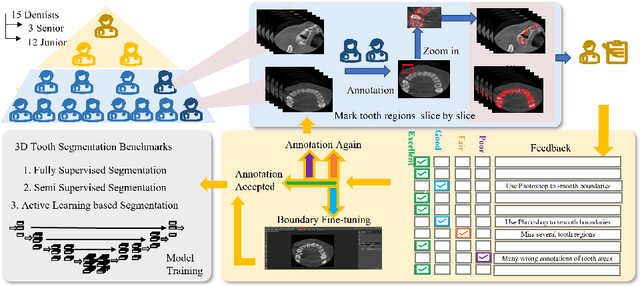
Abstract:Accurate tooth volume segmentation is a prerequisite for computer-aided dental analysis. Deep learning-based tooth segmentation methods have achieved satisfying performances but require a large quantity of tooth data with ground truth. The dental data publicly available is limited meaning the existing methods can not be reproduced, evaluated and applied in clinical practice. In this paper, we establish a 3D dental CBCT dataset CTooth+, with 22 fully annotated volumes and 146 unlabeled volumes. We further evaluate several state-of-the-art tooth volume segmentation strategies based on fully-supervised learning, semi-supervised learning and active learning, and define the performance principles. This work provides a new benchmark for the tooth volume segmentation task, and the experiment can serve as the baseline for future AI-based dental imaging research and clinical application development.
Attention Guided Network for Salient Object Detection in Optical Remote Sensing Images
Jul 05, 2022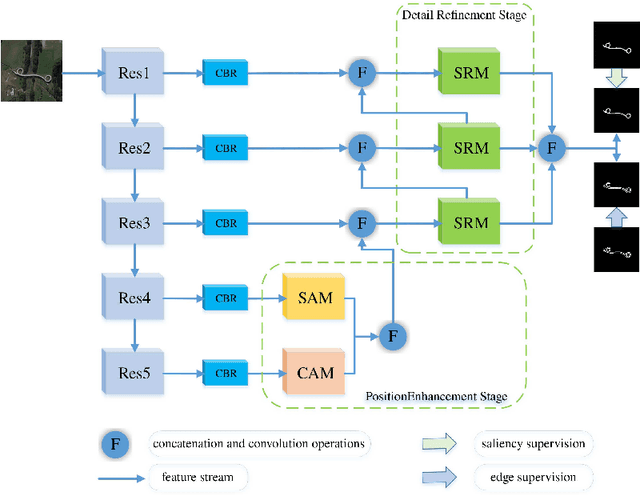
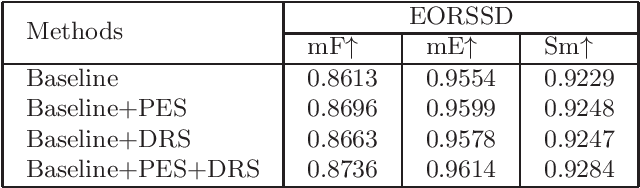
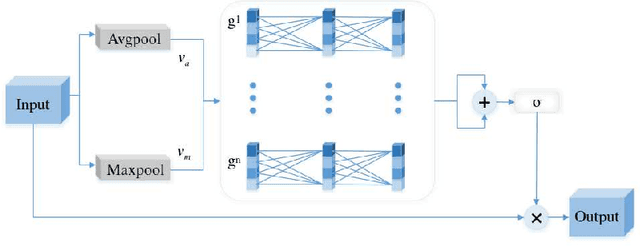
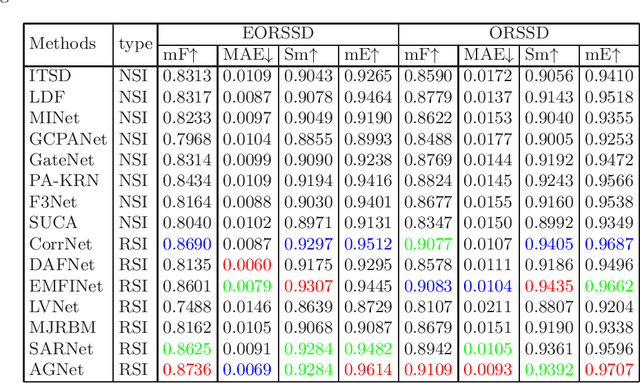
Abstract:Due to the extreme complexity of scale and shape as well as the uncertainty of the predicted location, salient object detection in optical remote sensing images (RSI-SOD) is a very difficult task. The existing SOD methods can satisfy the detection performance for natural scene images, but they are not well adapted to RSI-SOD due to the above-mentioned image characteristics in remote sensing images. In this paper, we propose a novel Attention Guided Network (AGNet) for SOD in optical RSIs, including position enhancement stage and detail refinement stage. Specifically, the position enhancement stage consists of a semantic attention module and a contextual attention module to accurately describe the approximate location of salient objects. The detail refinement stage uses the proposed self-refinement module to progressively refine the predicted results under the guidance of attention and reverse attention. In addition, the hybrid loss is applied to supervise the training of the network, which can improve the performance of the model from three perspectives of pixel, region and statistics. Extensive experiments on two popular benchmarks demonstrate that AGNet achieves competitive performance compared to other state-of-the-art methods. The code will be available at https://github.com/NuaaYH/AGNet.
DU-Net based Unsupervised Contrastive Learning for Cancer Segmentation in Histology Images
Jun 17, 2022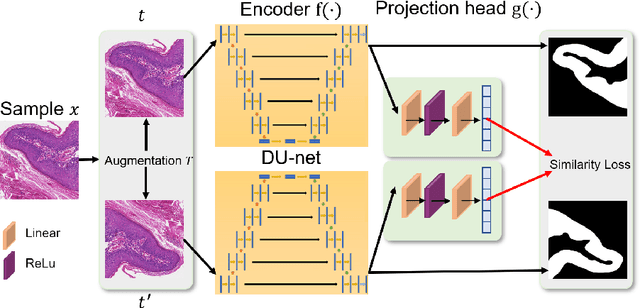
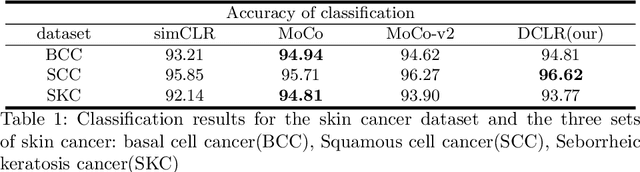
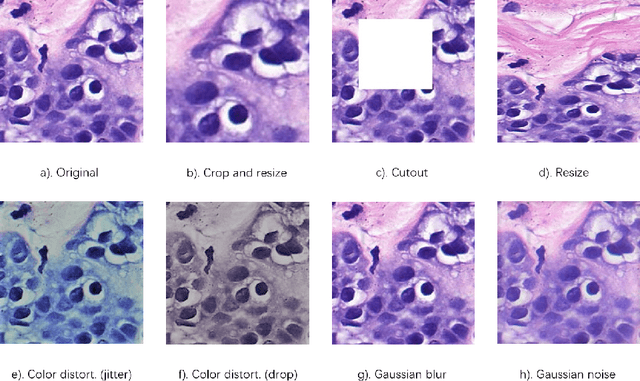
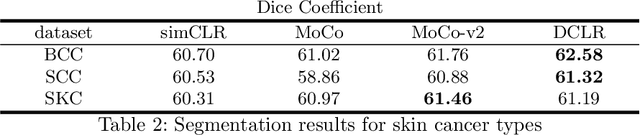
Abstract:In this paper, we introduce an unsupervised cancer segmentation framework for histology images. The framework involves an effective contrastive learning scheme for extracting distinctive visual representations for segmentation. The encoder is a Deep U-Net (DU-Net) structure that contains an extra fully convolution layer compared to the normal U-Net. A contrastive learning scheme is developed to solve the problem of lacking training sets with high-quality annotations on tumour boundaries. A specific set of data augmentation techniques are employed to improve the discriminability of the learned colour features from contrastive learning. Smoothing and noise elimination are conducted using convolutional Conditional Random Fields. The experiments demonstrate competitive performance in segmentation even better than some popular supervised networks.
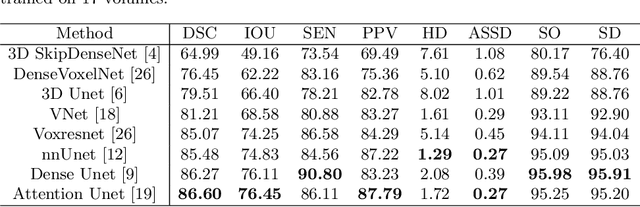
CTooth: A Fully Annotated 3D Dataset and Benchmark for Tooth Volume Segmentation on Cone Beam Computed Tomography Images
Jun 17, 2022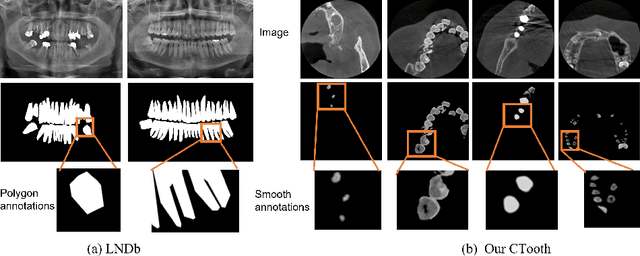
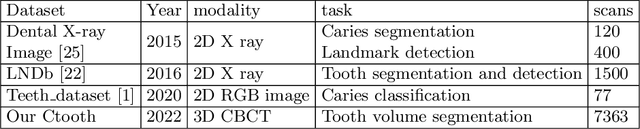
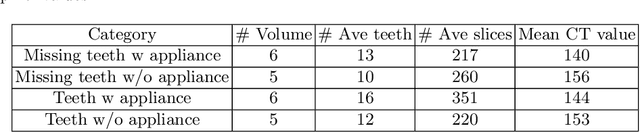
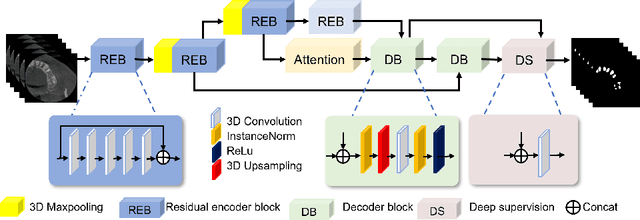
Abstract:3D tooth segmentation is a prerequisite for computer-aided dental diagnosis and treatment. However, segmenting all tooth regions manually is subjective and time-consuming. Recently, deep learning-based segmentation methods produce convincing results and reduce manual annotation efforts, but it requires a large quantity of ground truth for training. To our knowledge, there are few tooth data available for the 3D segmentation study. In this paper, we establish a fully annotated cone beam computed tomography dataset CTooth with tooth gold standard. This dataset contains 22 volumes (7363 slices) with fine tooth labels annotated by experienced radiographic interpreters. To ensure a relative even data sampling distribution, data variance is included in the CTooth including missing teeth and dental restoration. Several state-of-the-art segmentation methods are evaluated on this dataset. Afterwards, we further summarise and apply a series of 3D attention-based Unet variants for segmenting tooth volumes. This work provides a new benchmark for the tooth volume segmentation task. Experimental evidence proves that attention modules of the 3D UNet structure boost responses in tooth areas and inhibit the influence of background and noise. The best performance is achieved by 3D Unet with SKNet attention module, of 88.04 \% Dice and 78.71 \% IOU, respectively. The attention-based Unet framework outperforms other state-of-the-art methods on the CTooth dataset. The codebase and dataset are released.
Salient Skin Lesion Segmentation via Dilated Scale-Wise Feature Fusion Network
May 20, 2022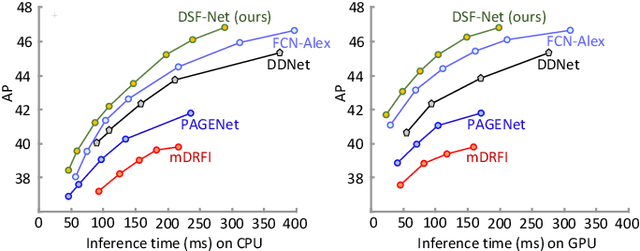
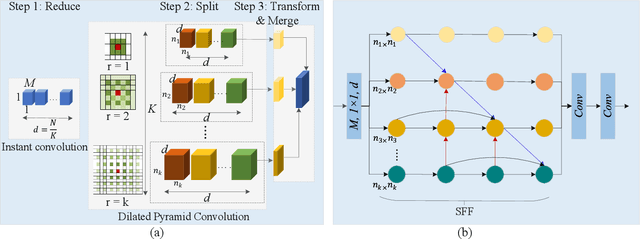
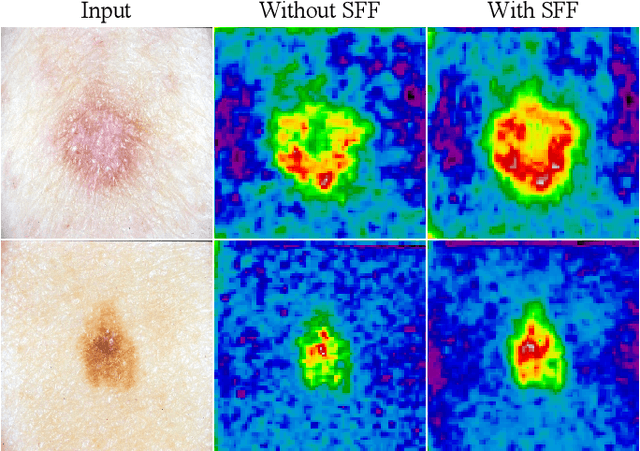
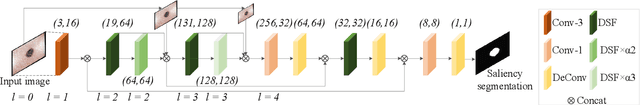
Abstract:Skin lesion detection in dermoscopic images is essential in the accurate and early diagnosis of skin cancer by a computerized apparatus. Current skin lesion segmentation approaches show poor performance in challenging circumstances such as indistinct lesion boundaries, low contrast between the lesion and the surrounding area, or heterogeneous background that causes over/under segmentation of the skin lesion. To accurately recognize the lesion from the neighboring regions, we propose a dilated scale-wise feature fusion network based on convolution factorization. Our network is designed to simultaneously extract features at different scales which are systematically fused for better detection. The proposed model has satisfactory accuracy and efficiency. Various experiments for lesion segmentation are performed along with comparisons with the state-of-the-art models. Our proposed model consistently showcases state-of-the-art results.
A lightweight multi-scale context network for salient object detection in optical remote sensing images
May 18, 2022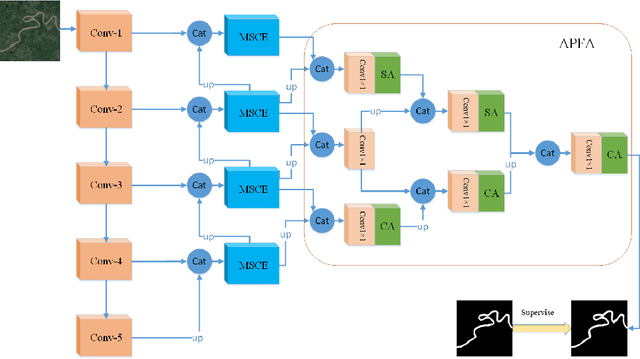
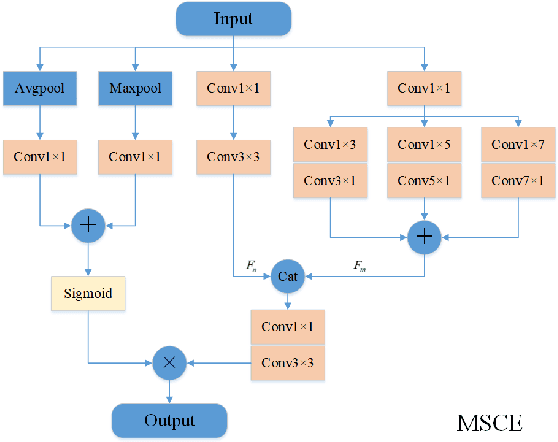
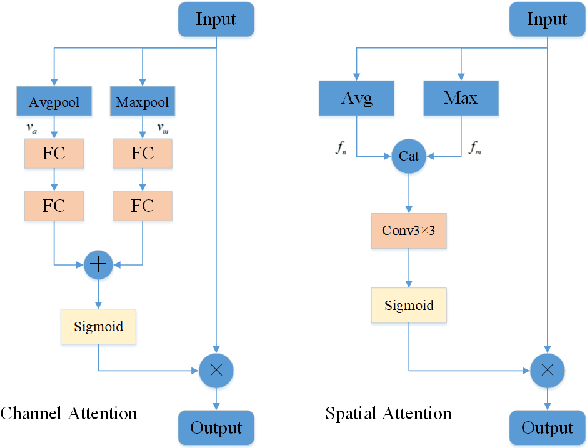

Abstract:Due to the more dramatic multi-scale variations and more complicated foregrounds and backgrounds in optical remote sensing images (RSIs), the salient object detection (SOD) for optical RSIs becomes a huge challenge. However, different from natural scene images (NSIs), the discussion on the optical RSI SOD task still remains scarce. In this paper, we propose a multi-scale context network, namely MSCNet, for SOD in optical RSIs. Specifically, a multi-scale context extraction module is adopted to address the scale variation of salient objects by effectively learning multi-scale contextual information. Meanwhile, in order to accurately detect complete salient objects in complex backgrounds, we design an attention-based pyramid feature aggregation mechanism for gradually aggregating and refining the salient regions from the multi-scale context extraction module. Extensive experiments on two benchmarks demonstrate that MSCNet achieves competitive performance with only 3.26M parameters. The code will be available at https://github.com/NuaaYH/MSCNet.
Absolute Wrong Makes Better: Boosting Weakly Supervised Object Detection via Negative Deterministic Information
Apr 21, 2022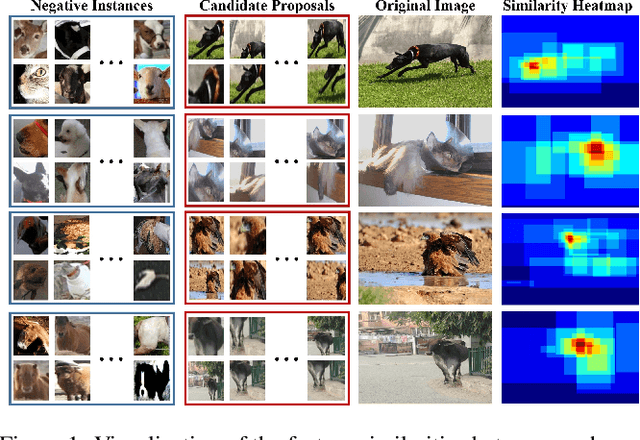
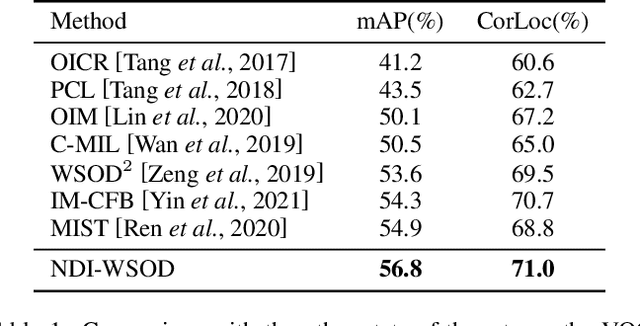
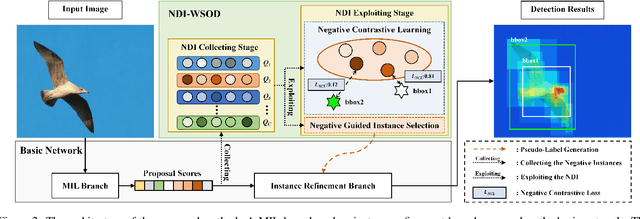
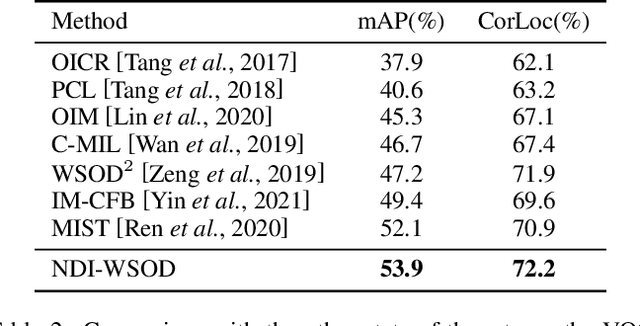
Abstract:Weakly supervised object detection (WSOD) is a challenging task, in which image-level labels (e.g., categories of the instances in the whole image) are used to train an object detector. Many existing methods follow the standard multiple instance learning (MIL) paradigm and have achieved promising performance. However, the lack of deterministic information leads to part domination and missing instances. To address these issues, this paper focuses on identifying and fully exploiting the deterministic information in WSOD. We discover that negative instances (i.e. absolutely wrong instances), ignored in most of the previous studies, normally contain valuable deterministic information. Based on this observation, we here propose a negative deterministic information (NDI) based method for improving WSOD, namely NDI-WSOD. Specifically, our method consists of two stages: NDI collecting and exploiting. In the collecting stage, we design several processes to identify and distill the NDI from negative instances online. In the exploiting stage, we utilize the extracted NDI to construct a novel negative contrastive learning mechanism and a negative guided instance selection strategy for dealing with the issues of part domination and missing instances, respectively. Experimental results on several public benchmarks including VOC 2007, VOC 2012 and MS COCO show that our method achieves satisfactory performance.
Source-free Domain Adaptation for Multi-site and Lifespan Brain Skull Stripping
Mar 11, 2022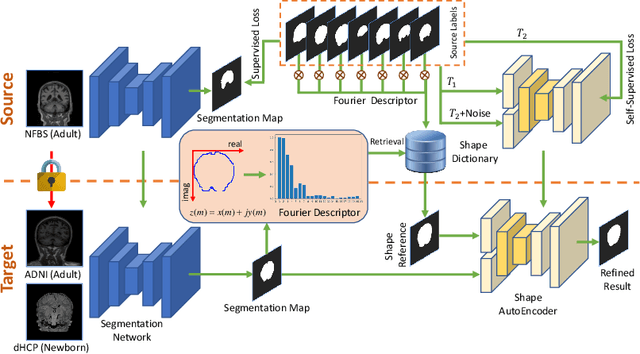
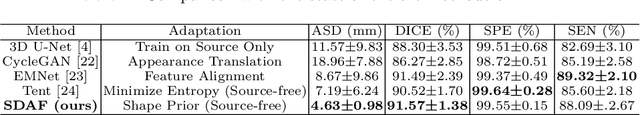
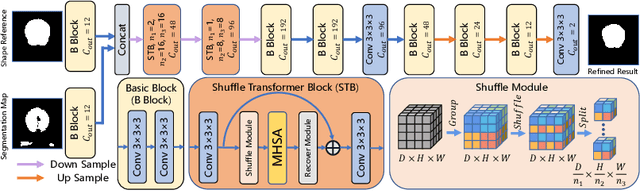
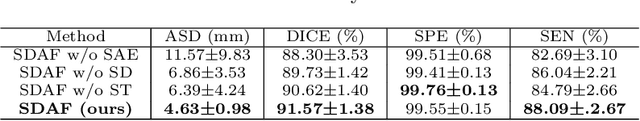
Abstract:Skull stripping is a crucial prerequisite step in the analysis of brain magnetic resonance (MR) images. Although many excellent works or tools have been proposed, they suffer from low generalization capability. For instance, the model trained on a dataset with specific imaging parameters (source domain) cannot be well applied to other datasets with different imaging parameters (target domain). Especially, for the lifespan datasets, the model trained on an adult dataset is not applicable to an infant dataset due to the large domain difference. To address this issue, numerous domain adaptation (DA) methods have been proposed to align the extracted features between the source and target domains, requiring concurrent access to the input images of both domains. Unfortunately, it is problematic to share the images due to privacy. In this paper, we design a source-free domain adaptation framework (SDAF) for multi-site and lifespan skull stripping that can accomplish domain adaptation without access to source domain images. Our method only needs to share the source labels as shape dictionaries and the weights trained on the source data, without disclosing private information from source domain subjects. To deal with the domain shift between multi-site lifespan datasets, we take advantage of the brain shape prior which is invariant to imaging parameters and ages. Experiments demonstrate that our framework can significantly outperform the state-of-the-art methods on multi-site lifespan datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge