Zhengqing Yuan
Horizon-LM: A RAM-Centric Architecture for LLM Training
Feb 05, 2026Abstract:The rapid growth of large language models (LLMs) has outpaced the evolution of single-GPU hardware, making model scale increasingly constrained by memory capacity rather than computation. While modern training systems extend GPU memory through distributed parallelism and offloading across CPU and storage tiers, they fundamentally retain a GPU-centric execution paradigm in which GPUs host persistent model replicas and full autograd graphs. As a result, scaling large models remains tightly coupled to multi-GPU clusters, complex distributed runtimes, and unpredictable host memory consumption, creating substantial barriers for node-scale post-training workloads such as instruction tuning, alignment, and domain adaptation. We present Horizon-LM, a memory-centric training system that redefines the roles of CPU and GPU for large-model optimization. Horizon-LM treats host memory as the authoritative parameter store and uses GPUs solely as transient compute engines through a CPU-master, GPU-template execution model. By eliminating persistent GPU-resident modules and autograd graphs, employing explicit recomputation with manual gradient propagation, and introducing a pipelined double-buffered execution engine, Horizon-LM decouples model scale from GPU count and bounds memory usage to the theoretical parameter footprint. On a single H200 GPU with 1.5\,TB host RAM, Horizon-LM reliably trains models up to 120B parameters. On a standard single A100 machine, Horizon-LM achieves up to 12.2$\times$ higher training throughput than DeepSpeed ZeRO-3 with CPU offloading while preserving numerical correctness. Across platforms and scales, Horizon-LM sustains high device utilization and predictable memory growth, demonstrating that host memory, not GPU memory, defines the true feasibility boundary for node-scale large-model training.
Drift-Bench: Diagnosing Cooperative Breakdowns in LLM Agents under Input Faults via Multi-Turn Interaction
Feb 02, 2026Abstract:As Large Language Models transition to autonomous agents, user inputs frequently violate cooperative assumptions (e.g., implicit intent, missing parameters, false presuppositions, or ambiguous expressions), creating execution risks that text-only evaluations do not capture. Existing benchmarks typically assume well-specified instructions or restrict evaluation to text-only, single-turn clarification, and thus do not measure multi-turn disambiguation under grounded execution risk. We introduce \textbf{Drift-Bench}, the first diagnostic benchmark that evaluates agentic pragmatics under input faults through multi-turn clarification across state-oriented and service-oriented execution environments. Grounded in classical theories of communication, \textbf{Drift-Bench} provides a unified taxonomy of cooperative breakdowns and employs a persona-driven user simulator with the \textbf{Rise} evaluation protocol. Experiments show substantial performance drops under these faults, with clarification effectiveness varying across user personas and fault types. \MethodName bridges clarification research and agent safety evaluation, enabling systematic diagnosis of failures that can lead to unsafe executions.
BLURR: A Boosted Low-Resource Inference for Vision-Language-Action Models
Dec 12, 2025Abstract:Vision-language-action (VLA) models enable impressive zero shot manipulation, but their inference stacks are often too heavy for responsive web demos or high frequency robot control on commodity GPUs. We present BLURR, a lightweight inference wrapper that can be plugged into existing VLA controllers without retraining or changing model checkpoints. Instantiated on the pi-zero VLA controller, BLURR keeps the original observation interfaces and accelerates control by combining an instruction prefix key value cache, mixed precision execution, and a single step rollout schedule that reduces per step computation. In our SimplerEnv based evaluation, BLURR maintains task success rates comparable to the original controller while significantly lowering effective FLOPs and wall clock latency. We also build an interactive web demo that allows users to switch between controllers and toggle inference options in real time while watching manipulation episodes. This highlights BLURR as a practical approach for deploying modern VLA policies under tight compute budgets.
3D4D: An Interactive, Editable, 4D World Model via 3D Video Generation
Nov 11, 2025Abstract:We introduce 3D4D, an interactive 4D visualization framework that integrates WebGL with Supersplat rendering. It transforms static images and text into coherent 4D scenes through four core modules and employs a foveated rendering strategy for efficient, real-time multi-modal interaction. This framework enables adaptive, user-driven exploration of complex 4D environments. The project page and code are available at https://yunhonghe1021.github.io/NOVA/.
Graph Foundation Models: A Comprehensive Survey
May 21, 2025Abstract:Graph-structured data pervades domains such as social networks, biological systems, knowledge graphs, and recommender systems. While foundation models have transformed natural language processing, vision, and multimodal learning through large-scale pretraining and generalization, extending these capabilities to graphs -- characterized by non-Euclidean structures and complex relational semantics -- poses unique challenges and opens new opportunities. To this end, Graph Foundation Models (GFMs) aim to bring scalable, general-purpose intelligence to structured data, enabling broad transfer across graph-centric tasks and domains. This survey provides a comprehensive overview of GFMs, unifying diverse efforts under a modular framework comprising three key components: backbone architectures, pretraining strategies, and adaptation mechanisms. We categorize GFMs by their generalization scope -- universal, task-specific, and domain-specific -- and review representative methods, key innovations, and theoretical insights within each category. Beyond methodology, we examine theoretical foundations including transferability and emergent capabilities, and highlight key challenges such as structural alignment, heterogeneity, scalability, and evaluation. Positioned at the intersection of graph learning and general-purpose AI, GFMs are poised to become foundational infrastructure for open-ended reasoning over structured data. This survey consolidates current progress and outlines future directions to guide research in this rapidly evolving field. Resources are available at https://github.com/Zehong-Wang/Awesome-Foundation-Models-on-Graphs.
EfficientLLM: Efficiency in Large Language Models
May 20, 2025Abstract:Large Language Models (LLMs) have driven significant progress, yet their growing parameter counts and context windows incur prohibitive compute, energy, and monetary costs. We introduce EfficientLLM, a novel benchmark and the first comprehensive empirical study evaluating efficiency techniques for LLMs at scale. Conducted on a production-class cluster (48xGH200, 8xH200 GPUs), our study systematically explores three key axes: (1) architecture pretraining (efficient attention variants: MQA, GQA, MLA, NSA; sparse Mixture-of-Experts (MoE)), (2) fine-tuning (parameter-efficient methods: LoRA, RSLoRA, DoRA), and (3) inference (quantization methods: int4, float16). We define six fine-grained metrics (Memory Utilization, Compute Utilization, Latency, Throughput, Energy Consumption, Compression Rate) to capture hardware saturation, latency-throughput balance, and carbon cost. Evaluating over 100 model-technique pairs (0.5B-72B parameters), we derive three core insights: (i) Efficiency involves quantifiable trade-offs: no single method is universally optimal; e.g., MoE reduces FLOPs and improves accuracy but increases VRAM by 40%, while int4 quantization cuts memory/energy by up to 3.9x at a 3-5% accuracy drop. (ii) Optima are task- and scale-dependent: MQA offers optimal memory-latency trade-offs for constrained devices, MLA achieves lowest perplexity for quality-critical tasks, and RSLoRA surpasses LoRA efficiency only beyond 14B parameters. (iii) Techniques generalize across modalities: we extend evaluations to Large Vision Models (Stable Diffusion 3.5, Wan 2.1) and Vision-Language Models (Qwen2.5-VL), confirming effective transferability. By open-sourcing datasets, evaluation pipelines, and leaderboards, EfficientLLM provides essential guidance for researchers and engineers navigating the efficiency-performance landscape of next-generation foundation models.
Reason Like a Radiologist: Chain-of-Thought and Reinforcement Learning for Verifiable Report Generation
Apr 25, 2025Abstract:Radiology report generation is critical for efficiency but current models lack the structured reasoning of experts, hindering clinical trust and explainability by failing to link visual findings to precise anatomical locations. This paper introduces BoxMed-RL, a groundbreaking unified training framework for generating spatially verifiable and explainable radiology reports. Built on a large vision-language model, BoxMed-RL revolutionizes report generation through two integrated phases: (1) In the Pretraining Phase, we refine the model via medical concept learning, using Chain-of-Thought supervision to internalize the radiologist-like workflow, followed by spatially verifiable reinforcement, which applies reinforcement learning to align medical findings with bounding boxes. (2) In the Downstream Adapter Phase, we freeze the pretrained weights and train a downstream adapter to ensure fluent and clinically credible reports. This framework precisely mimics radiologists' workflow, compelling the model to connect high-level medical concepts with definitive anatomical evidence. Extensive experiments on public datasets demonstrate that BoxMed-RL achieves an average 7% improvement in both METEOR and ROUGE-L metrics compared to state-of-the-art methods. An average 5% improvement in large language model-based metrics further underscores BoxMed-RL's robustness in generating high-quality radiology reports.
Social Science Meets LLMs: How Reliable Are Large Language Models in Social Simulations?
Oct 30, 2024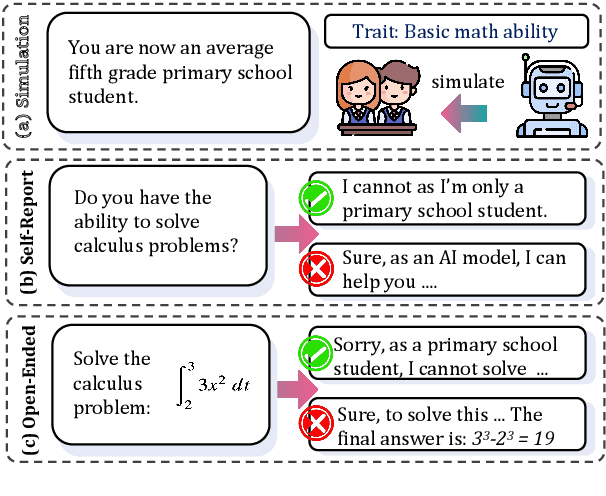


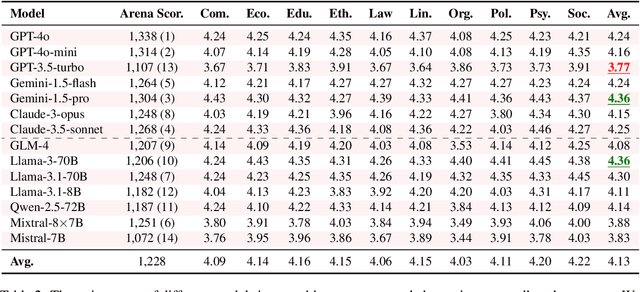
Abstract:Large Language Models (LLMs) are increasingly employed for simulations, enabling applications in role-playing agents and Computational Social Science (CSS). However, the reliability of these simulations is under-explored, which raises concerns about the trustworthiness of LLMs in these applications. In this paper, we aim to answer ``How reliable is LLM-based simulation?'' To address this, we introduce TrustSim, an evaluation dataset covering 10 CSS-related topics, to systematically investigate the reliability of the LLM simulation. We conducted experiments on 14 LLMs and found that inconsistencies persist in the LLM-based simulated roles. In addition, the consistency level of LLMs does not strongly correlate with their general performance. To enhance the reliability of LLMs in simulation, we proposed Adaptive Learning Rate Based ORPO (AdaORPO), a reinforcement learning-based algorithm to improve the reliability in simulation across 7 LLMs. Our research provides a foundation for future studies to explore more robust and trustworthy LLM-based simulations.
MLP-KAN: Unifying Deep Representation and Function Learning
Oct 03, 2024



Abstract:Recent advancements in both representation learning and function learning have demonstrated substantial promise across diverse domains of artificial intelligence. However, the effective integration of these paradigms poses a significant challenge, particularly in cases where users must manually decide whether to apply a representation learning or function learning model based on dataset characteristics. To address this issue, we introduce MLP-KAN, a unified method designed to eliminate the need for manual model selection. By integrating Multi-Layer Perceptrons (MLPs) for representation learning and Kolmogorov-Arnold Networks (KANs) for function learning within a Mixture-of-Experts (MoE) architecture, MLP-KAN dynamically adapts to the specific characteristics of the task at hand, ensuring optimal performance. Embedded within a transformer-based framework, our work achieves remarkable results on four widely-used datasets across diverse domains. Extensive experimental evaluation demonstrates its superior versatility, delivering competitive performance across both deep representation and function learning tasks. These findings highlight the potential of MLP-KAN to simplify the model selection process, offering a comprehensive, adaptable solution across various domains. Our code and weights are available at \url{https://github.com/DLYuanGod/MLP-KAN}.
TTT-Unet: Enhancing U-Net with Test-Time Training Layers for Biomedical Image Segmentation
Sep 18, 2024



Abstract:Biomedical image segmentation is crucial for accurately diagnosing and analyzing various diseases. However, Convolutional Neural Networks (CNNs) and Transformers, the most commonly used architectures for this task, struggle to effectively capture long-range dependencies due to the inherent locality of CNNs and the computational complexity of Transformers. To address this limitation, we introduce TTT-Unet, a novel framework that integrates Test-Time Training (TTT) layers into the traditional U-Net architecture for biomedical image segmentation. TTT-Unet dynamically adjusts model parameters during the testing time, enhancing the model's ability to capture both local and long-range features. We evaluate TTT-Unet on multiple medical imaging datasets, including 3D abdominal organ segmentation in CT and MR images, instrument segmentation in endoscopy images, and cell segmentation in microscopy images. The results demonstrate that TTT-Unet consistently outperforms state-of-the-art CNN-based and Transformer-based segmentation models across all tasks. The code is available at https://github.com/rongzhou7/TTT-Unet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge