Hui Ren
MedReason: Eliciting Factual Medical Reasoning Steps in LLMs via Knowledge Graphs
Apr 01, 2025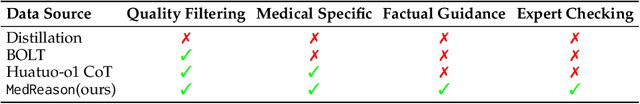
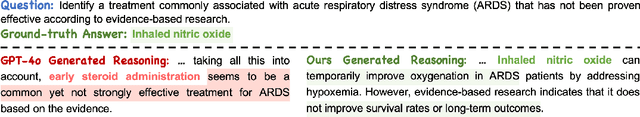
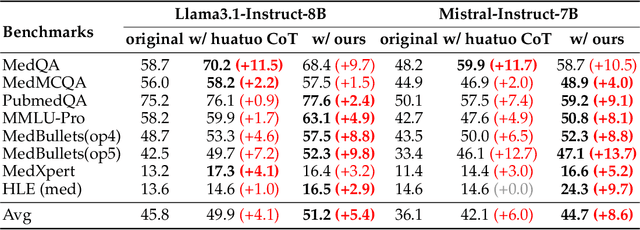
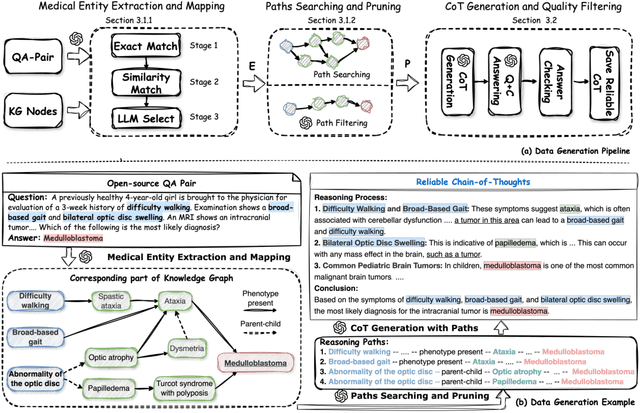
Abstract:Medical tasks such as diagnosis and treatment planning require precise and complex reasoning, particularly in life-critical domains. Unlike mathematical reasoning, medical reasoning demands meticulous, verifiable thought processes to ensure reliability and accuracy. However, there is a notable lack of datasets that provide transparent, step-by-step reasoning to validate and enhance the medical reasoning ability of AI models. To bridge this gap, we introduce MedReason, a large-scale high-quality medical reasoning dataset designed to enable faithful and explainable medical problem-solving in large language models (LLMs). We utilize a structured medical knowledge graph (KG) to convert clinical QA pairs into logical chains of reasoning, or ``thinking paths'', which trace connections from question elements to answers via relevant KG entities. Each path is validated for consistency with clinical logic and evidence-based medicine. Our pipeline generates detailed reasoning for various medical questions from 7 medical datasets, resulting in a dataset of 32,682 question-answer pairs, each with detailed, step-by-step explanations. Experiments demonstrate that fine-tuning with our dataset consistently boosts medical problem-solving capabilities, achieving significant gains of up to 7.7% for DeepSeek-Ditill-8B. Our top-performing model, MedReason-8B, outperforms the Huatuo-o1-8B, a state-of-the-art medical reasoning model, by up to 4.2% on the clinical benchmark MedBullets. We also engage medical professionals from diverse specialties to assess our dataset's quality, ensuring MedReason offers accurate and coherent medical reasoning. Our data, models, and code will be publicly available.
Feature4X: Bridging Any Monocular Video to 4D Agentic AI with Versatile Gaussian Feature Fields
Mar 26, 2025Abstract:Recent advancements in 2D and multimodal models have achieved remarkable success by leveraging large-scale training on extensive datasets. However, extending these achievements to enable free-form interactions and high-level semantic operations with complex 3D/4D scenes remains challenging. This difficulty stems from the limited availability of large-scale, annotated 3D/4D or multi-view datasets, which are crucial for generalizable vision and language tasks such as open-vocabulary and prompt-based segmentation, language-guided editing, and visual question answering (VQA). In this paper, we introduce Feature4X, a universal framework designed to extend any functionality from 2D vision foundation model into the 4D realm, using only monocular video input, which is widely available from user-generated content. The "X" in Feature4X represents its versatility, enabling any task through adaptable, model-conditioned 4D feature field distillation. At the core of our framework is a dynamic optimization strategy that unifies multiple model capabilities into a single representation. Additionally, to the best of our knowledge, Feature4X is the first method to distill and lift the features of video foundation models (e.g. SAM2, InternVideo2) into an explicit 4D feature field using Gaussian Splatting. Our experiments showcase novel view segment anything, geometric and appearance scene editing, and free-form VQA across all time steps, empowered by LLMs in feedback loops. These advancements broaden the scope of agentic AI applications by providing a foundation for scalable, contextually and spatiotemporally aware systems capable of immersive dynamic 4D scene interaction.
Art-Free Generative Models: Art Creation Without Graphic Art Knowledge
Nov 29, 2024



Abstract:We explore the question: "How much prior art knowledge is needed to create art?" To investigate this, we propose a text-to-image generation model trained without access to art-related content. We then introduce a simple yet effective method to learn an art adapter using only a few examples of selected artistic styles. Our experiments show that art generated using our method is perceived by users as comparable to art produced by models trained on large, art-rich datasets. Finally, through data attribution techniques, we illustrate how examples from both artistic and non-artistic datasets contributed to the creation of new artistic styles.
EchoFM: Foundation Model for Generalizable Echocardiogram Analysis
Oct 30, 2024



Abstract:Foundation models have recently gained significant attention because of their generalizability and adaptability across multiple tasks and data distributions. Although medical foundation models have emerged, solutions for cardiac imaging, especially echocardiography videos, are still unexplored. In this paper, we introduce EchoFM, a foundation model specifically designed to represent and analyze echocardiography videos. In EchoFM, we propose a self-supervised learning framework that captures both spatial and temporal variability patterns through a spatio-temporal consistent masking strategy and periodic-driven contrastive learning. This framework can effectively capture the spatio-temporal dynamics of echocardiography and learn the representative video features without any labels. We pre-train our model on an extensive dataset comprising over 290,000 echocardiography videos covering 26 scan views across different imaging modes, with up to 20 million frames of images. The pre-trained EchoFM can then be easily adapted and fine-tuned for a variety of downstream tasks, serving as a robust backbone model. Our evaluation was systemically designed for four downstream tasks after the echocardiography examination routine. Experiment results show that EchoFM surpasses state-of-the-art methods, including specialized echocardiography methods, self-supervised pre-training models, and general-purposed pre-trained foundation models, across all downstream tasks.
Dual-level Adaptive Self-Labeling for Novel Class Discovery in Point Cloud Segmentation
Jul 17, 2024



Abstract:We tackle the novel class discovery in point cloud segmentation, which discovers novel classes based on the semantic knowledge of seen classes. Existing work proposes an online point-wise clustering method with a simplified equal class-size constraint on the novel classes to avoid degenerate solutions. However, the inherent imbalanced distribution of novel classes in point clouds typically violates the equal class-size constraint. Moreover, point-wise clustering ignores the rich spatial context information of objects, which results in less expressive representation for semantic segmentation. To address the above challenges, we propose a novel self-labeling strategy that adaptively generates high-quality pseudo-labels for imbalanced classes during model training. In addition, we develop a dual-level representation that incorporates regional consistency into the point-level classifier learning, reducing noise in generated segmentation. Finally, we conduct extensive experiments on two widely used datasets, SemanticKITTI and SemanticPOSS, and the results show our method outperforms the state of the art by a large margin.
Biomedical Visual Instruction Tuning with Clinician Preference Alignment
Jun 19, 2024



Abstract:Recent advancements in multimodal foundation models have showcased impressive capabilities in understanding and reasoning with visual and textual information. Adapting these foundation models trained for general usage to specialized domains like biomedicine requires large-scale domain-specific instruction datasets. While existing works have explored curating such datasets automatically, the resultant datasets are not explicitly aligned with domain expertise. In this work, we propose a data-centric framework, Biomedical Visual Instruction Tuning with Clinician Preference Alignment (BioMed-VITAL), that incorporates clinician preferences into both stages of generating and selecting instruction data for tuning biomedical multimodal foundation models. First, during the generation stage, we prompt the GPT-4V generator with a diverse set of clinician-selected demonstrations for preference-aligned data candidate generation. Then, during the selection phase, we train a separate selection model, which explicitly distills clinician and policy-guided model preferences into a rating function to select high-quality data for medical instruction tuning. Results show that the model tuned with the instruction-following data from our method demonstrates a significant improvement in open visual chat (18.5% relatively) and medical VQA (win rate up to 81.73%). Our instruction-following data and models are available at BioMed-VITAL.github.io.
Prompt-driven Universal Model for View-Agnostic Echocardiography Analysis
Apr 09, 2024Abstract:Echocardiography segmentation for cardiac analysis is time-consuming and resource-intensive due to the variability in image quality and the necessity to process scans from various standard views. While current automated segmentation methods in echocardiography show promising performance, they are trained on specific scan views to analyze corresponding data. However, this solution has a limitation as the number of required models increases with the number of standard views. To address this, in this paper, we present a prompt-driven universal method for view-agnostic echocardiography analysis. Considering the domain shift between standard views, we first introduce a method called prompt matching, aimed at learning prompts specific to different views by matching prompts and querying input embeddings using a pre-trained vision model. Then, we utilized a pre-trained medical language model to align textual information with pixel data for accurate segmentation. Extensive experiments on three standard views showed that our approach significantly outperforms the state-of-the-art universal methods and achieves comparable or even better performances over the segmentation model trained and tested on same views.
SP$^2$OT: Semantic-Regularized Progressive Partial Optimal Transport for Imbalanced Clustering
Apr 04, 2024Abstract:Deep clustering, which learns representation and semantic clustering without labels information, poses a great challenge for deep learning-based approaches. Despite significant progress in recent years, most existing methods focus on uniformly distributed datasets, significantly limiting the practical applicability of their methods. In this paper, we propose a more practical problem setting named deep imbalanced clustering, where the underlying classes exhibit an imbalance distribution. To address this challenge, we introduce a novel optimal transport-based pseudo-label learning framework. Our framework formulates pseudo-label generation as a Semantic-regularized Progressive Partial Optimal Transport (SP$^2$OT) problem, which progressively transports each sample to imbalanced clusters under several prior distribution and semantic relation constraints, thus generating high-quality and imbalance-aware pseudo-labels. To solve SP$^2$OT, we develop a Majorization-Minimization-based optimization algorithm. To be more precise, we employ the strategy of majorization to reformulate the SP$^2$OT problem into a Progressive Partial Optimal Transport problem, which can be transformed into an unbalanced optimal transport problem with augmented constraints and can be solved efficiently by a fast matrix scaling algorithm. Experiments on various datasets, including a human-curated long-tailed CIFAR100, challenging ImageNet-R, and large-scale subsets of fine-grained iNaturalist2018 datasets, demonstrate the superiority of our method.
Cardiac Magnetic Resonance 2D+T Short- and Long-axis Segmentation via Spatio-temporal SAM Adaptation
Mar 15, 2024Abstract:Accurate 2D+T myocardium segmentation in cine cardiac magnetic resonance (CMR) scans is essential to analyze LV motion throughout the cardiac cycle comprehensively. The Segment Anything Model (SAM), known for its accurate segmentation and zero-shot generalization, has not yet been tailored for CMR 2D+T segmentation. We therefore introduce CMR2D+T-SAM, a novel approach to adapt SAM for CMR 2D+T segmentation using spatio-temporal adaption. This approach also incorporates a U-Net framework for multi-scale feature extraction, as well as text prompts for accurate segmentation on both short-axis (SAX) and long-axis (LAX) views using a single model. CMR2D+T-SAM outperforms existing deep learning methods on the STACOM2011 dataset, achieving a myocardium Dice score of 0.885 and a Hausdorff distance (HD) of 2.900 pixels. It also demonstrates superior zero-shot generalization on the ACDC dataset with a Dice score of 0.840 and a HD of 4.076 pixels.
Conditional Score-Based Diffusion Model for Cortical Thickness Trajectory Prediction
Mar 11, 2024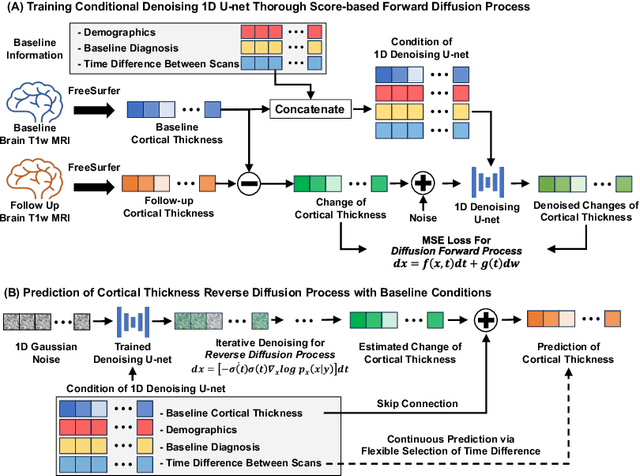
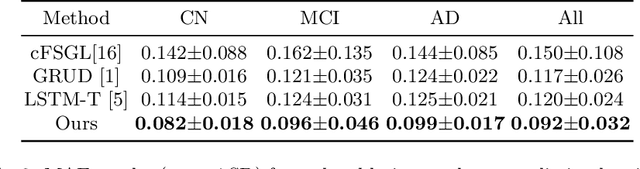
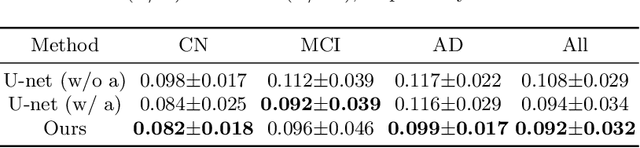
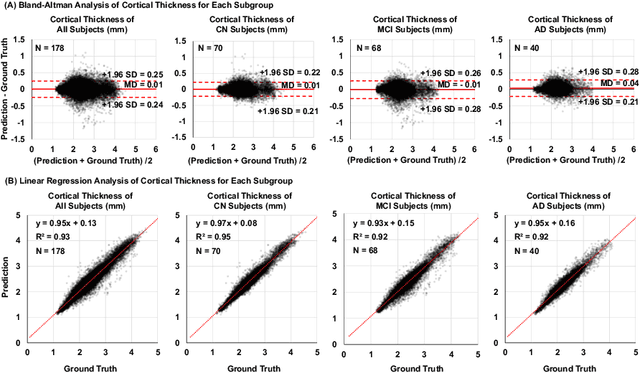
Abstract:Alzheimer's Disease (AD) is a neurodegenerative condition characterized by diverse progression rates among individuals, with changes in cortical thickness (CTh) closely linked to its progression. Accurately forecasting CTh trajectories can significantly enhance early diagnosis and intervention strategies, providing timely care. However, the longitudinal data essential for these studies often suffer from temporal sparsity and incompleteness, presenting substantial challenges in modeling the disease's progression accurately. Existing methods are limited, focusing primarily on datasets without missing entries or requiring predefined assumptions about CTh progression. To overcome these obstacles, we propose a conditional score-based diffusion model specifically designed to generate CTh trajectories with the given baseline information, such as age, sex, and initial diagnosis. Our conditional diffusion model utilizes all available data during the training phase to make predictions based solely on baseline information during inference without needing prior history about CTh progression. The prediction accuracy of the proposed CTh prediction pipeline using a conditional score-based model was compared for sub-groups consisting of cognitively normal, mild cognitive impairment, and AD subjects. The Bland-Altman analysis shows our diffusion-based prediction model has a near-zero bias with narrow 95% confidential interval compared to the ground-truth CTh in 6-36 months. In addition, our conditional diffusion model has a stochastic generative nature, therefore, we demonstrated an uncertainty analysis of patient-specific CTh prediction through multiple realizations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge