Universal Lesion Detection by Learning from Multiple Heterogeneously Labeled Datasets
May 28, 2020Ke Yan, Jinzheng Cai, Adam P. Harrison, Dakai Jin, Jing Xiao, Le Lu
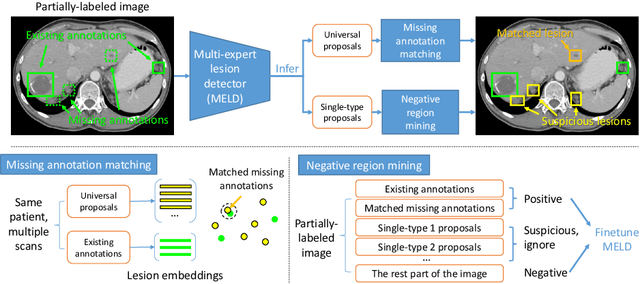
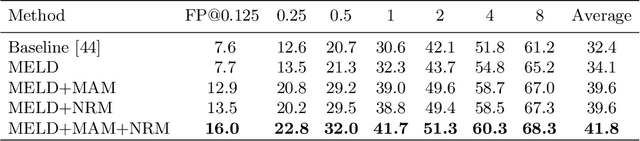
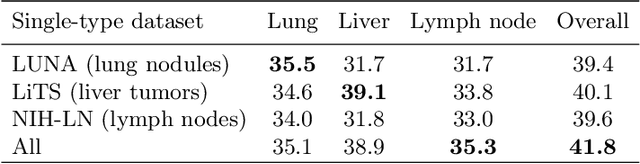
Lesion detection is an important problem within medical imaging analysis. Most previous work focuses on detecting and segmenting a specialized category of lesions (e.g., lung nodules). However, in clinical practice, radiologists are responsible for finding all possible types of anomalies. The task of universal lesion detection (ULD) was proposed to address this challenge by detecting a large variety of lesions from the whole body. There are multiple heterogeneously labeled datasets with varying label completeness: DeepLesion, the largest dataset of 32,735 annotated lesions of various types, but with even more missing annotation instances; and several fully-labeled single-type lesion datasets, such as LUNA for lung nodules and LiTS for liver tumors. In this work, we propose a novel framework to leverage all these datasets together to improve the performance of ULD. First, we learn a multi-head multi-task lesion detector using all datasets and generate lesion proposals on DeepLesion. Second, missing annotations in DeepLesion are retrieved by a new method of embedding matching that exploits clinical prior knowledge. Last, we discover suspicious but unannotated lesions using knowledge transfer from single-type lesion detectors. In this way, reliable positive and negative regions are obtained from partially-labeled and unlabeled images, which are effectively utilized to train ULD. To assess the clinically realistic protocol of 3D volumetric ULD, we fully annotated 1071 CT sub-volumes in DeepLesion. Our method outperforms the current state-of-the-art approach by 29% in the metric of average sensitivity.
Detecting Scatteredly-Distributed, Small, andCritically Important Objects in 3D OncologyImaging via Decision Stratification
May 27, 2020Zhuotun Zhu, Ke Yan, Dakai Jin, Jinzheng Cai, Tsung-Ying Ho, Adam P Harrison, Dazhou Guo, Chun-Hung Chao, Xianghua Ye, Jing Xiao, Alan Yuille, Le Lu
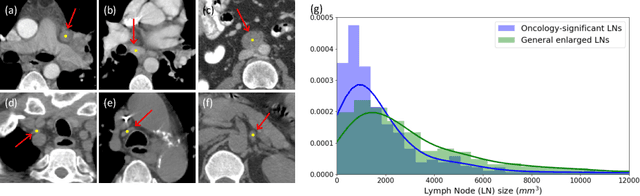
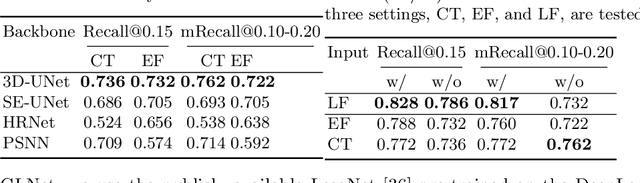
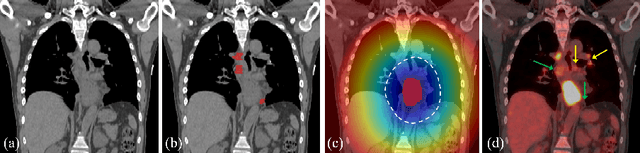
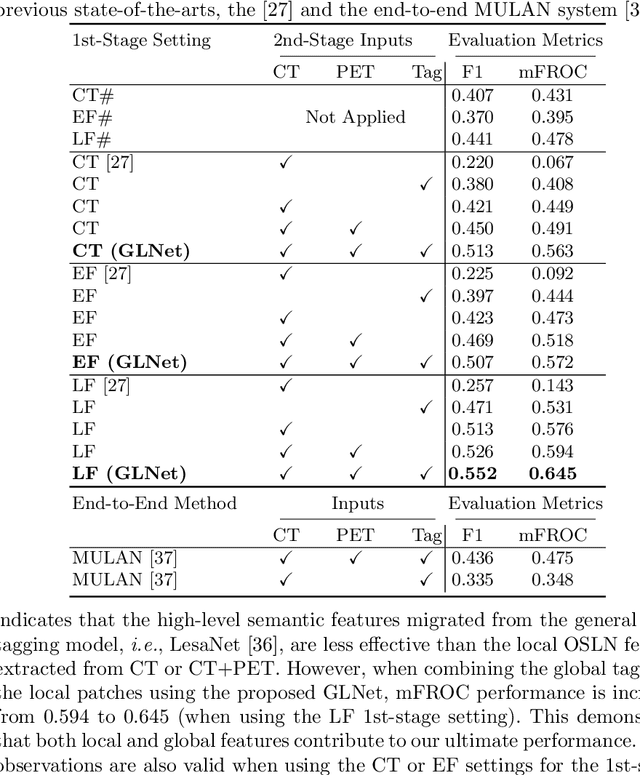
Finding and identifying scatteredly-distributed, small, and critically important objects in 3D oncology images is very challenging. We focus on the detection and segmentation of oncology-significant (or suspicious cancer metastasized) lymph nodes (OSLNs), which has not been studied before as a computational task. Determining and delineating the spread of OSLNs is essential in defining the corresponding resection/irradiating regions for the downstream workflows of surgical resection and radiotherapy of various cancers. For patients who are treated with radiotherapy, this task is performed by experienced radiation oncologists that involves high-level reasoning on whether LNs are metastasized, which is subject to high inter-observer variations. In this work, we propose a divide-and-conquer decision stratification approach that divides OSLNs into tumor-proximal and tumor-distal categories. This is motivated by the observation that each category has its own different underlying distributions in appearance, size and other characteristics. Two separate detection-by-segmentation networks are trained per category and fused. To further reduce false positives (FP), we present a novel global-local network (GLNet) that combines high-level lesion characteristics with features learned from localized 3D image patches. Our method is evaluated on a dataset of 141 esophageal cancer patients with PET and CT modalities (the largest to-date). Our results significantly improve the recall from $45\%$ to $67\%$ at $3$ FPs per patient as compared to previous state-of-the-art methods. The highest achieved OSLN recall of $0.828$ is clinically relevant and valuable.
JSSR: A Joint Synthesis, Segmentation, and Registration System for 3D Multi-Modal Image Alignment of Large-scale Pathological CT Scans
May 27, 2020Fengze Liu, Jingzheng Cai, Yuankai Huo, Chi-Tung Cheng, Ashwin Raju, Dakai Jin, Jing Xiao, Alan Yuille, Le Lu, ChienHung Liao, Adam P Harrison
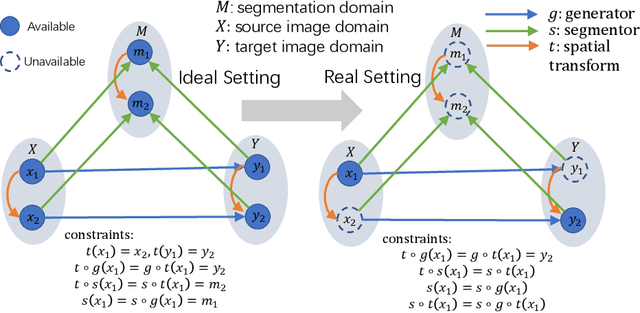
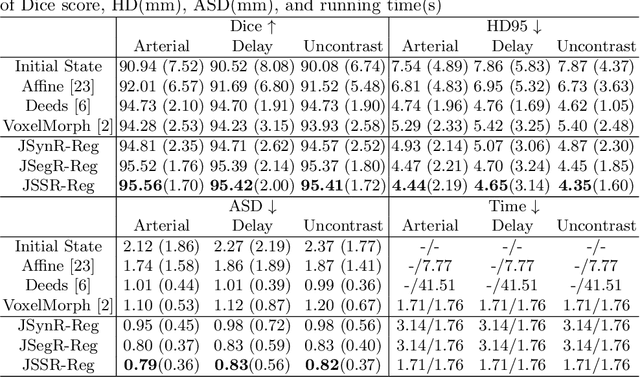
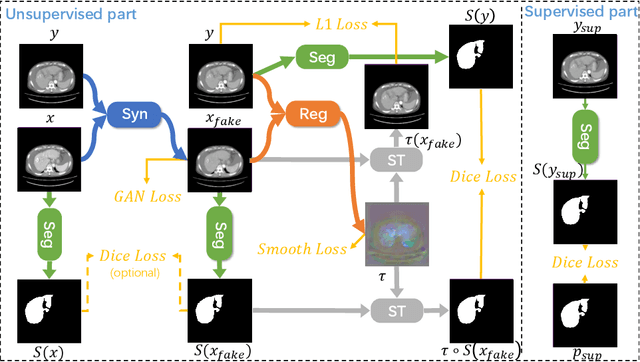
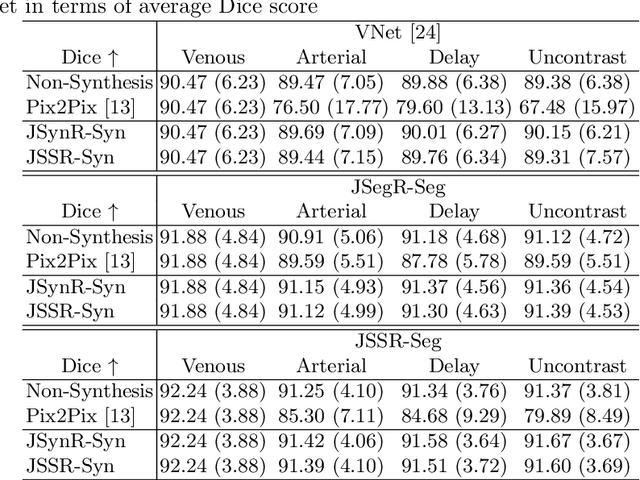
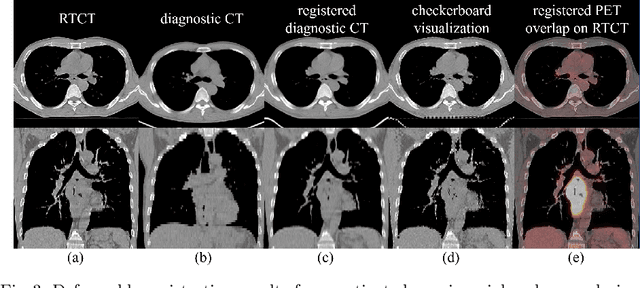
Multi-modal image registration is a challenging problem yet important clinical task in many real applications and scenarios. For medical imaging based diagnosis, deformable registration among different image modalities is often required in order to provide complementary visual information, as the first step. During the registration, the semantic information is the key to match homologous points and pixels. Nevertheless, many conventional registration methods are incapable to capture the high-level semantic anatomical dense correspondences. In this work, we propose a novel multi-task learning system, JSSR, based on an end-to-end 3D convolutional neural network that is composed of a generator, a register and a segmentor, for the tasks of synthesis, registration and segmentation, respectively. This system is optimized to satisfy the implicit constraints between different tasks unsupervisedly. It first synthesizes the source domain images into the target domain, then an intra-modal registration is applied on the synthesized images and target images. Then we can get the semantic segmentation by applying segmentors on the synthesized images and target images, which are aligned by the same deformation field generated by the registers. The supervision from another fully-annotated dataset is used to regularize the segmentors. We extensively evaluate our JSSR system on a large-scale medical image dataset containing 1,485 patient CT imaging studies of four different phases (i.e., 5,940 3D CT scans with pathological livers) on the registration, segmentation and synthesis tasks. The performance is improved after joint training on the registration and segmentation tasks by 0.9% and 1.9% respectively from a highly competitive and accurate baseline. The registration part also consistently outperforms the conventional state-of-the-art multi-modal registration methods.
Organ at Risk Segmentation for Head and Neck Cancer using Stratified Learning and Neural Architecture Search
Apr 17, 2020Dazhou Guo, Dakai Jin, Zhuotun Zhu, Tsung-Ying Ho, Adam P. Harrison, Chun-Hung Chao, Jing Xiao, Alan Yuille, Chien-Yu Lin, Le Lu
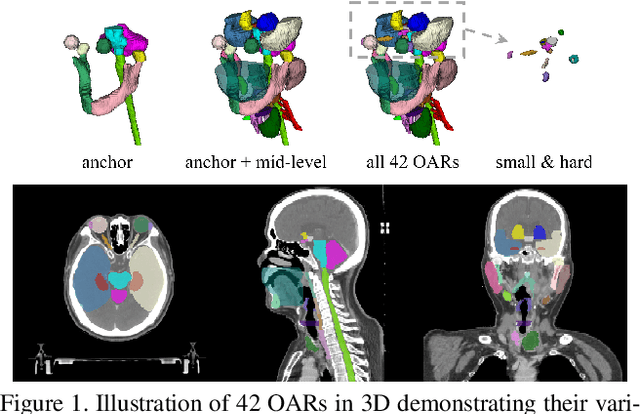
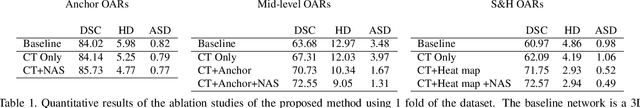
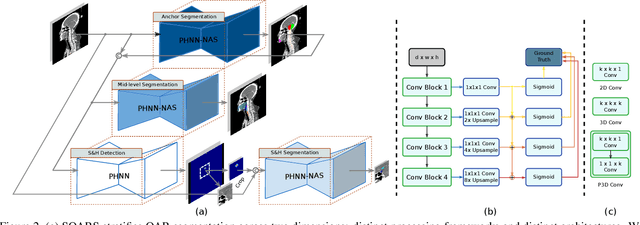
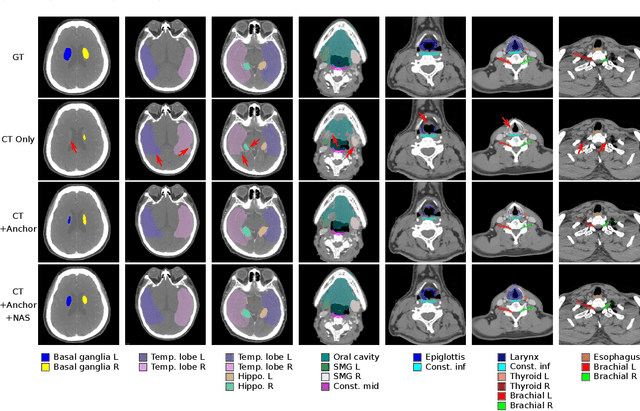
OAR segmentation is a critical step in radiotherapy of head and neck (H&N) cancer, where inconsistencies across radiation oncologists and prohibitive labor costs motivate automated approaches. However, leading methods using standard fully convolutional network workflows that are challenged when the number of OARs becomes large, e.g. > 40. For such scenarios, insights can be gained from the stratification approaches seen in manual clinical OAR delineation. This is the goal of our work, where we introduce stratified organ at risk segmentation (SOARS), an approach that stratifies OARs into anchor, mid-level, and small & hard (S&H) categories. SOARS stratifies across two dimensions. The first dimension is that distinct processing pipelines are used for each OAR category. In particular, inspired by clinical practices, anchor OARs are used to guide the mid-level and S&H categories. The second dimension is that distinct network architectures are used to manage the significant contrast, size, and anatomy variations between different OARs. We use differentiable neural architecture search (NAS), allowing the network to choose among 2D, 3D or Pseudo-3D convolutions. Extensive 4-fold cross-validation on 142 H&N cancer patients with 42 manually labeled OARs, the most comprehensive OAR dataset to date, demonstrates that both pipeline- and NAS-stratification significantly improves quantitative performance over the state-of-the-art (from 69.52% to 73.68% in absolute Dice scores). Thus, SOARS provides a powerful and principled means to manage the highly complex segmentation space of OARs.
Deep Esophageal Clinical Target Volume Delineation using Encoded 3D Spatial Context of Tumors, Lymph Nodes, and Organs At Risk
Sep 06, 2019Dakai Jin, Dazhou Guo, Tsung-Ying Ho, Adam P. Harrison, Jing Xiao, Chen-kan Tseng, Le Lu
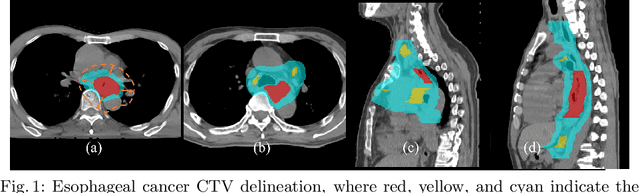

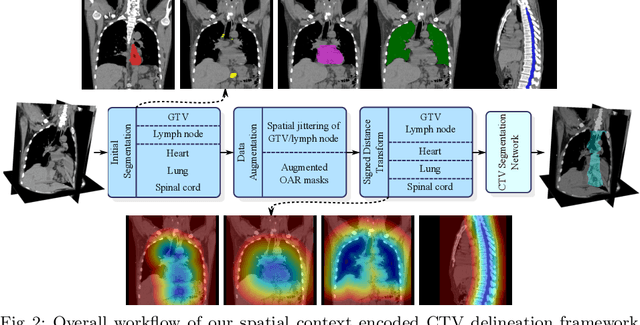
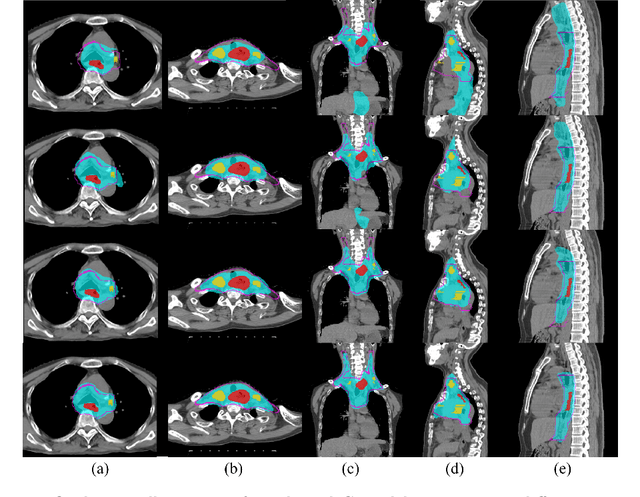
Clinical target volume (CTV) delineation from radiotherapy computed tomography (RTCT) images is used to define the treatment areas containing the gross tumor volume (GTV) and/or sub-clinical malignant disease for radiotherapy (RT). High intra- and inter-user variability makes this a particularly difficult task for esophageal cancer. This motivates automated solutions, which is the aim of our work. Because CTV delineation is highly context-dependent--it must encompass the GTV and regional lymph nodes (LNs) while also avoiding excessive exposure to the organs at risk (OARs)--we formulate it as a deep contextual appearance-based problem using encoded spatial contexts of these anatomical structures. This allows the deep network to better learn from and emulate the margin- and appearance-based delineation performed by human physicians. Additionally, we develop domain-specific data augmentation to inject robustness to our system. Finally, we show that a simple 3D progressive holistically nested network (PHNN), which avoids computationally heavy decoding paths while still aggregating features at different levels of context, can outperform more complicated networks. Cross-validated experiments on a dataset of 135 esophageal cancer patients demonstrate that our encoded spatial context approach can produce concrete performance improvements, with an average Dice score of 83.9% and an average surface distance of 4.2 mm, representing improvements of 3.8% and 2.4 mm, respectively, over the state-of-the-art approach.
Accurate Esophageal Gross Tumor Volume Segmentation in PET/CT using Two-Stream Chained 3D Deep Network Fusion
Sep 06, 2019Dakai Jin, Dazhou Guo, Tsung-Ying Ho, Adam P. Harrison, Jing Xiao, Chen-kan Tseng, Le Lu
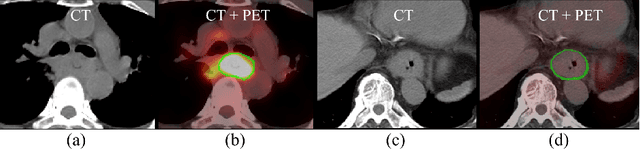
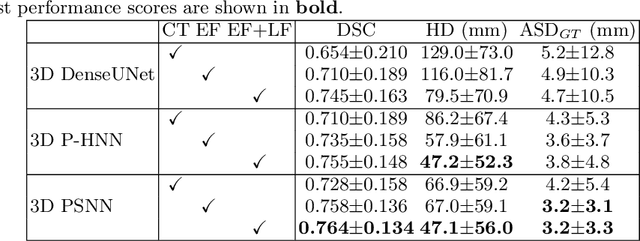
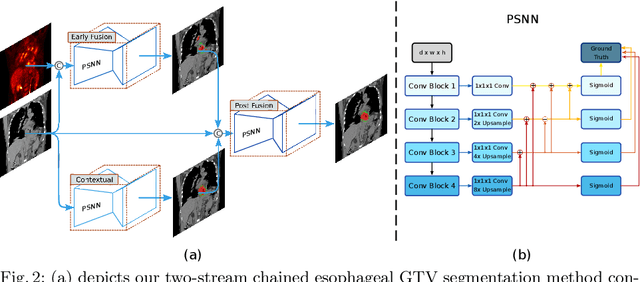
Gross tumor volume (GTV) segmentation is a critical step in esophageal cancer radiotherapy treatment planning. Inconsistencies across oncologists and prohibitive labor costs motivate automated approaches for this task. However, leading approaches are only applied to radiotherapy computed tomography (RTCT) images taken prior to treatment. This limits the performance as RTCT suffers from low contrast between the esophagus, tumor, and surrounding tissues. In this paper, we aim to exploit both RTCT and positron emission tomography (PET) imaging modalities to facilitate more accurate GTV segmentation. By utilizing PET, we emulate medical professionals who frequently delineate GTV boundaries through observation of the RTCT images obtained after prescribing radiotherapy and PET/CT images acquired earlier for cancer staging. To take advantage of both modalities, we present a two-stream chained segmentation approach that effectively fuses the CT and PET modalities via early and late 3D deep-network-based fusion. Furthermore, to effect the fusion and segmentation we propose a simple yet effective progressive semantically nested network (PSNN) model that outperforms more complicated models. Extensive 5-fold cross-validation on 110 esophageal cancer patients, the largest analysis to date, demonstrates that both the proposed two-stream chained segmentation pipeline and the PSNN model can significantly improve the quantitative performance over the previous state-of-the-art work by 11% in absolute Dice score (DSC) (from 0.654 to 0.764) and, at the same time, reducing the Hausdorff distance from 129 mm to 47 mm.
Weakly Supervised Universal Fracture Detection in Pelvic X-rays
Sep 04, 2019Yirui Wang, Le Lu, Chi-Tung Cheng, Dakai Jin, Adam P. Harrison, Jing Xiao, Chien-Hung Liao, Shun Miao
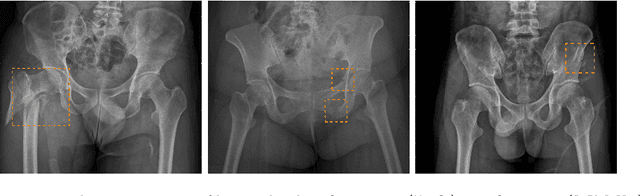
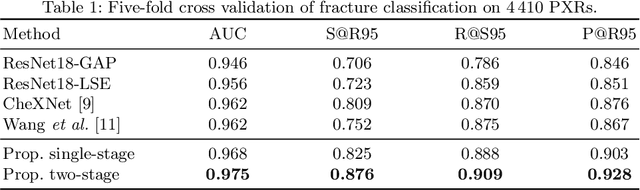
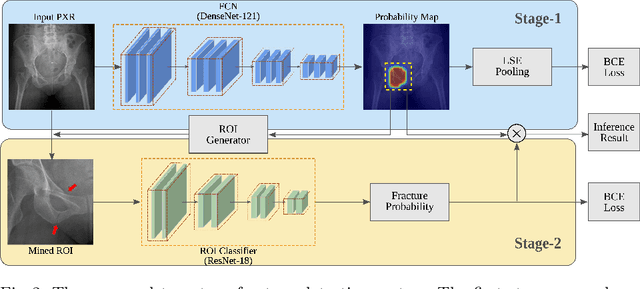
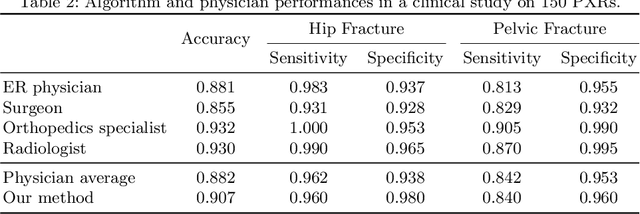
Hip and pelvic fractures are serious injuries with life-threatening complications. However, diagnostic errors of fractures in pelvic X-rays (PXRs) are very common, driving the demand for computer-aided diagnosis (CAD) solutions. A major challenge lies in the fact that fractures are localized patterns that require localized analyses. Unfortunately, the PXRs residing in hospital picture archiving and communication system do not typically specify region of interests. In this paper, we propose a two-stage hip and pelvic fracture detection method that executes localized fracture classification using weakly supervised ROI mining. The first stage uses a large capacity fully-convolutional network, i.e., deep with high levels of abstraction, in a multiple instance learning setting to automatically mine probable true positive and definite hard negative ROIs from the whole PXR in the training data. The second stage trains a smaller capacity model, i.e., shallower and more generalizable, with the mined ROIs to perform localized analyses to classify fractures. During inference, our method detects hip and pelvic fractures in one pass by chaining the probability outputs of the two stages together. We evaluate our method on 4 410 PXRs, reporting an area under the ROC curve value of 0.975, the highest among state-of-the-art fracture detection methods. Moreover, we show that our two-stage approach can perform comparably to human physicians (even outperforming emergency physicians and surgeons), in a preliminary reader study of 23 readers.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019Hugo J. Kuijf, J. Matthijs Biesbroek, Jeroen de Bresser, Rutger Heinen, Simon Andermatt, Mariana Bento, Matt Berseth, Mikhail Belyaev, M. Jorge Cardoso, Adrià Casamitjana, D. Louis Collins, Mahsa Dadar, Achilleas Georgiou, Mohsen Ghafoorian, Dakai Jin, April Khademi, Jesse Knight, Hongwei Li, Xavier Lladó, Miguel Luna, Qaiser Mahmood, Richard McKinley, Alireza Mehrtash, Sébastien Ourselin, Bo-yong Park, Hyunjin Park, Sang Hyun Park, Simon Pezold, Elodie Puybareau, Leticia Rittner, Carole H. Sudre, Sergi Valverde, Verónica Vilaplana, Roland Wiest, Yongchao Xu, Ziyue Xu, Guodong Zeng, Jianguo Zhang, Guoyan Zheng, Christopher Chen, Wiesje van der Flier, Frederik Barkhof, Max A. Viergever, Geert Jan Biessels
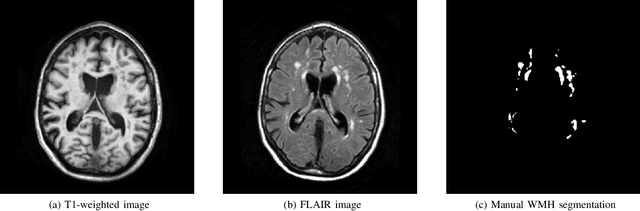
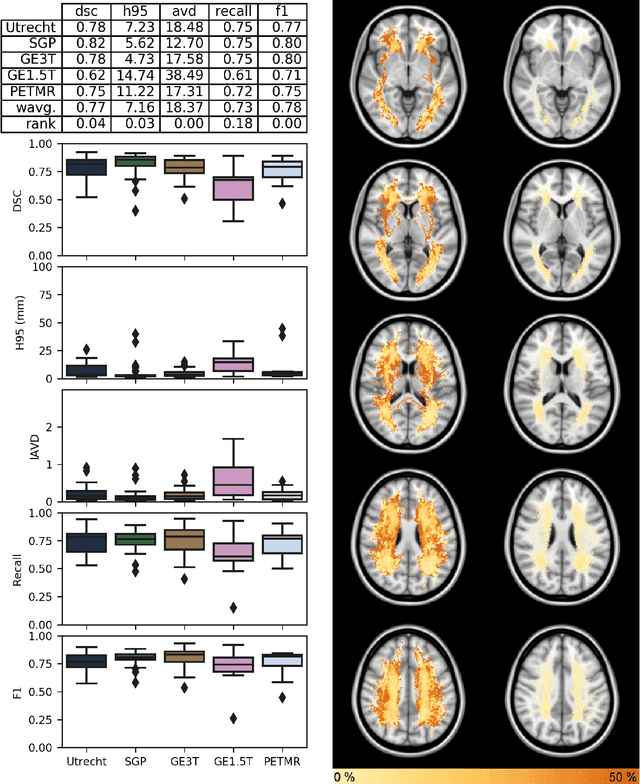
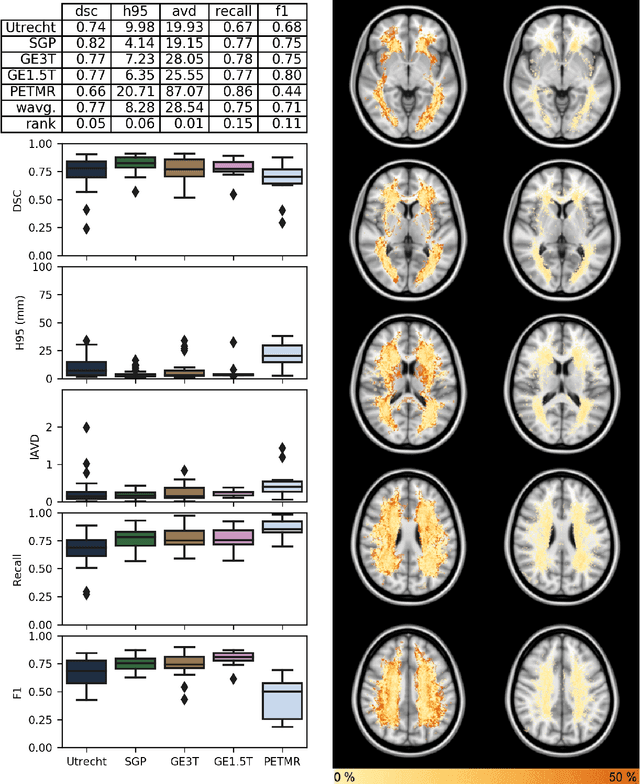
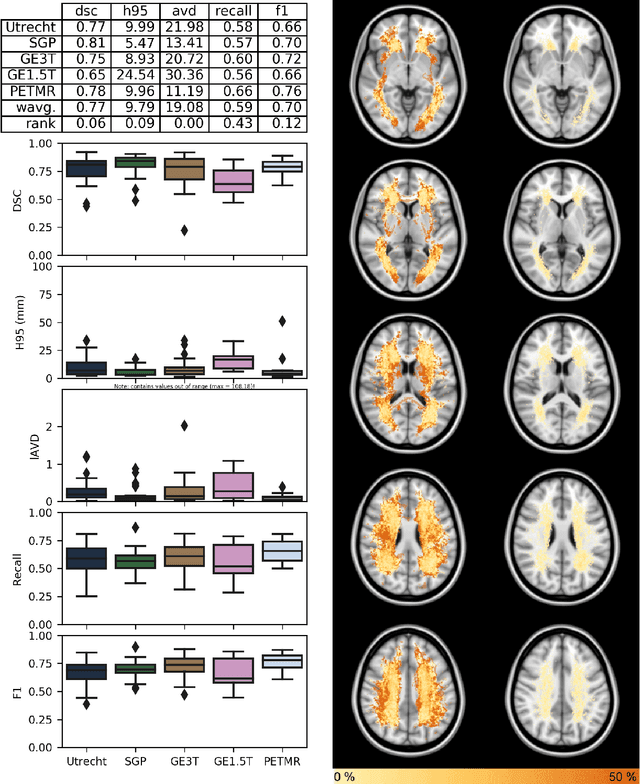
Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge