Hyunjin Park
Privacy-Preserving Chest X-ray Classification in Latent Space with Homomorphically Encrypted Neural Inference
Jun 18, 2025



Abstract:Medical imaging data contain sensitive patient information requiring strong privacy protection. Many analytical setups require data to be sent to a server for inference purposes. Homomorphic encryption (HE) provides a solution by allowing computations to be performed on encrypted data without revealing the original information. However, HE inference is computationally expensive, particularly for large images (e.g., chest X-rays). In this study, we propose an HE inference framework for medical images that uses VQGAN to compress images into latent representations, thereby significantly reducing the computational burden while preserving image quality. We approximate the activation functions with lower-degree polynomials to balance the accuracy and efficiency in compliance with HE requirements. We observed that a downsampling factor of eight for compression achieved an optimal balance between performance and computational cost. We further adapted the squeeze and excitation module, which is known to improve traditional CNNs, to enhance the HE framework. Our method was tested on two chest X-ray datasets for multi-label classification tasks using vanilla CNN backbones. Although HE inference remains relatively slow and introduces minor performance differences compared with unencrypted inference, our approach shows strong potential for practical use in medical images
Domain Aware Multi-Task Pretraining of 3D Swin Transformer for T1-weighted Brain MRI
Oct 01, 2024



Abstract:The scarcity of annotated medical images is a major bottleneck in developing learning models for medical image analysis. Hence, recent studies have focused on pretrained models with fewer annotation requirements that can be fine-tuned for various downstream tasks. However, existing approaches are mainly 3D adaptions of 2D approaches ill-suited for 3D medical imaging data. Motivated by this gap, we propose novel domain-aware multi-task learning tasks to pretrain a 3D Swin Transformer for brain magnetic resonance imaging (MRI). Our method considers the domain knowledge in brain MRI by incorporating brain anatomy and morphology as well as standard pretext tasks adapted for 3D imaging in a contrastive learning setting. We pretrain our model using large-scale brain MRI data of 13,687 samples spanning several large-scale databases. Our method outperforms existing supervised and self-supervised methods in three downstream tasks of Alzheimer's disease classification, Parkinson's disease classification, and age prediction tasks. The ablation study of the proposed pretext tasks shows the effectiveness of our pretext tasks.
RadiomicsFill-Mammo: Synthetic Mammogram Mass Manipulation with Radiomics Features
Jul 08, 2024Abstract:Motivated by the question, "Can we generate tumors with desired attributes?'' this study leverages radiomics features to explore the feasibility of generating synthetic tumor images. Characterized by its low-dimensional yet biologically meaningful markers, radiomics bridges the gap between complex medical imaging data and actionable clinical insights. We present RadiomicsFill-Mammo, the first of the RadiomicsFill series, an innovative technique that generates realistic mammogram mass images mirroring specific radiomics attributes using masked images and opposite breast images, leveraging a recent stable diffusion model. This approach also allows for the incorporation of essential clinical variables, such as BI-RADS and breast density, alongside radiomics features as conditions for mass generation. Results indicate that RadiomicsFill-Mammo effectively generates diverse and realistic tumor images based on various radiomics conditions. Results also demonstrate a significant improvement in mass detection capabilities, leveraging RadiomicsFill-Mammo as a strategy to generate simulated samples. Furthermore, RadiomicsFill-Mammo not only advances medical imaging research but also opens new avenues for enhancing treatment planning and tumor simulation. Our code is available at https://github.com/nainye/RadiomicsFill.
Radiomics-guided Multimodal Self-attention Network for Predicting Pathological Complete Response in Breast MRI
Jun 05, 2024Abstract:Breast cancer is the most prevalent cancer among women and predicting pathologic complete response (pCR) after anti-cancer treatment is crucial for patient prognosis and treatment customization. Deep learning has shown promise in medical imaging diagnosis, particularly when utilizing multiple imaging modalities to enhance accuracy. This study presents a model that predicts pCR in breast cancer patients using dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) and apparent diffusion coefficient (ADC) maps. Radiomics features are established hand-crafted features of the tumor region and thus could be useful in medical image analysis. Our approach extracts features from both DCE MRI and ADC using an encoder with a self-attention mechanism, leveraging radiomics to guide feature extraction from tumor-related regions. Our experimental results demonstrate the superior performance of our model in predicting pCR compared to other baseline methods.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Synthetic Tumor Manipulation: With Radiomics Features
Nov 05, 2023Abstract:We introduce RadiomicsFill, a synthetic tumor generator conditioned on radiomics features, enabling detailed control and individual manipulation of tumor subregions. This conditioning leverages conventional high-dimensional features of the tumor (i.e., radiomics features) and thus is biologically well-grounded. Our model combines generative adversarial networks, radiomics-feature conditioning, and multi-task learning. Through experiments with glioma patients, RadiomicsFill demonstrated its capability to generate diverse, realistic tumors and its fine-tuning ability for specific radiomics features like 'Pixel Surface' and 'Shape Sphericity'. The ability of RadiomicsFill to generate an unlimited number of realistic synthetic tumors offers notable prospects for both advancing medical imaging research and potential clinical applications.
Adaptive Latent Diffusion Model for 3D Medical Image to Image Translation: Multi-modal Magnetic Resonance Imaging Study
Nov 01, 2023



Abstract:Multi-modal images play a crucial role in comprehensive evaluations in medical image analysis providing complementary information for identifying clinically important biomarkers. However, in clinical practice, acquiring multiple modalities can be challenging due to reasons such as scan cost, limited scan time, and safety considerations. In this paper, we propose a model based on the latent diffusion model (LDM) that leverages switchable blocks for image-to-image translation in 3D medical images without patch cropping. The 3D LDM combined with conditioning using the target modality allows generating high-quality target modality in 3D overcoming the shortcoming of the missing out-of-slice information in 2D generation methods. The switchable block, noted as multiple switchable spatially adaptive normalization (MS-SPADE), dynamically transforms source latents to the desired style of the target latents to help with the diffusion process. The MS-SPADE block allows us to have one single model to tackle many translation tasks of one source modality to various targets removing the need for many translation models for different scenarios. Our model exhibited successful image synthesis across different source-target modality scenarios and surpassed other models in quantitative evaluations tested on multi-modal brain magnetic resonance imaging datasets of four different modalities and an independent IXI dataset. Our model demonstrated successful image synthesis across various modalities even allowing for one-to-many modality translations. Furthermore, it outperformed other one-to-one translation models in quantitative evaluations.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019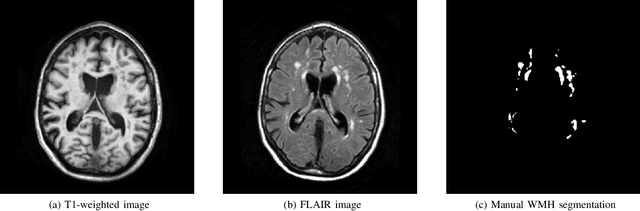
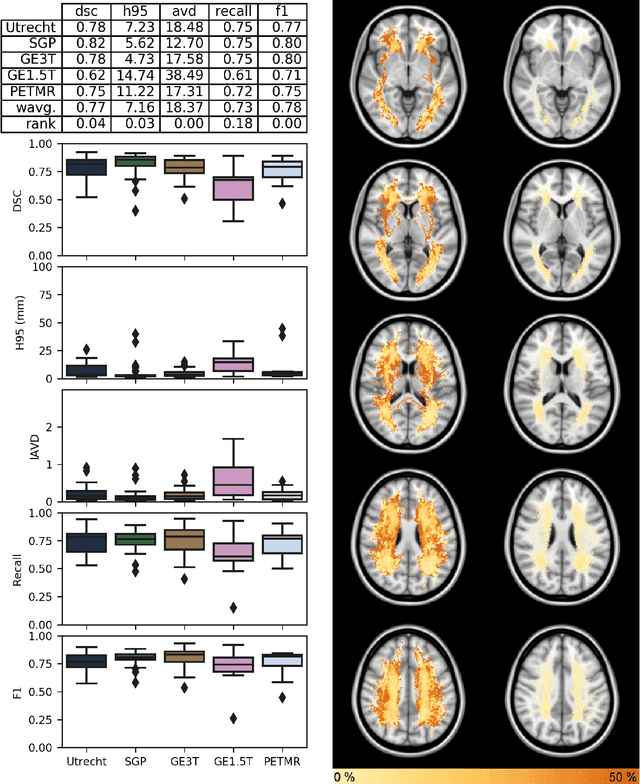
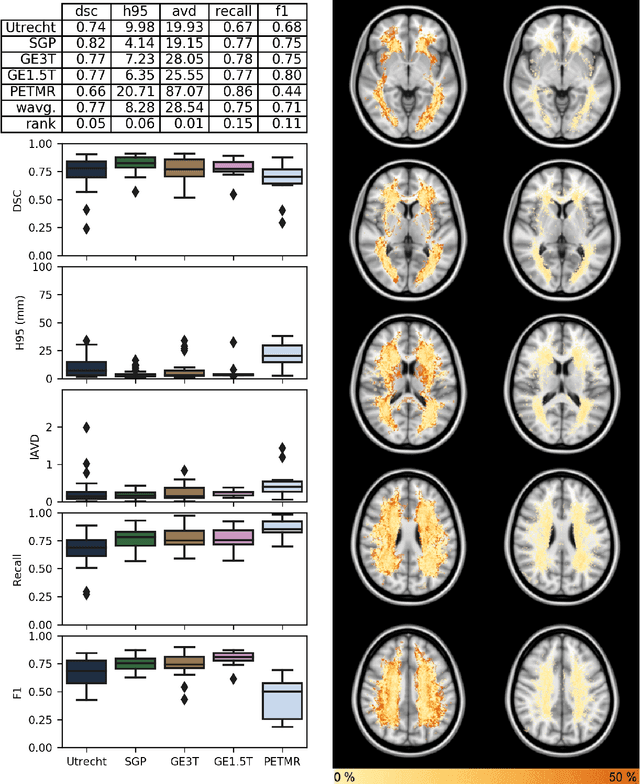
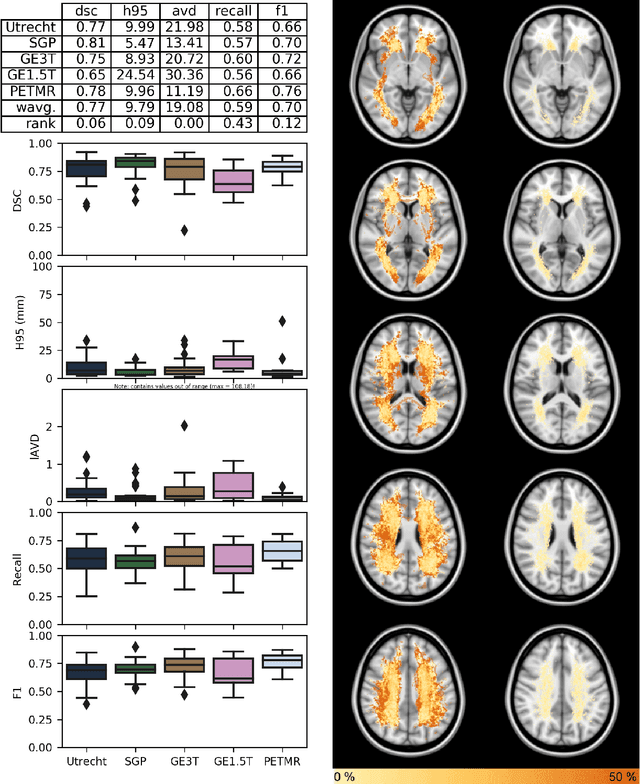
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge