Frederik Barkhof
for the ALFA study
LLM enhanced graph inference for long-term disease progression modelling
Nov 14, 2025Abstract:Understanding the interactions between biomarkers among brain regions during neurodegenerative disease is essential for unravelling the mechanisms underlying disease progression. For example, pathophysiological models of Alzheimer's Disease (AD) typically describe how variables, such as regional levels of toxic proteins, interact spatiotemporally within a dynamical system driven by an underlying biological substrate, often based on brain connectivity. However, current methods grossly oversimplify the complex relationship between brain connectivity by assuming a single-modality brain connectome as the disease-spreading substrate. This leads to inaccurate predictions of pathology spread, especially during the long-term progression period. Meanhwile, other methods of learning such a graph in a purely data-driven way face the identifiability issue due to lack of proper constraint. We thus present a novel framework that uses Large Language Models (LLMs) as expert guides on the interaction of regional variables to enhance learning of disease progression from irregularly sampled longitudinal patient data. By leveraging LLMs' ability to synthesize multi-modal relationships and incorporate diverse disease-driving mechanisms, our method simultaneously optimizes 1) the construction of long-term disease trajectories from individual-level observations and 2) the biologically-constrained graph structure that captures interactions among brain regions with better identifiability. We demonstrate the new approach by estimating the pathology propagation using tau-PET imaging data from an Alzheimer's disease cohort. The new framework demonstrates superior prediction accuracy and interpretability compared to traditional approaches while revealing additional disease-driving factors beyond conventional connectivity measures.
Automatic and standardized surgical reporting for central nervous system tumors
Aug 12, 2025Abstract:Magnetic resonance (MR) imaging is essential for evaluating central nervous system (CNS) tumors, guiding surgical planning, treatment decisions, and assessing postoperative outcomes and complication risks. While recent work has advanced automated tumor segmentation and report generation, most efforts have focused on preoperative data, with limited attention to postoperative imaging analysis. This study introduces a comprehensive pipeline for standardized postsurtical reporting in CNS tumors. Using the Attention U-Net architecture, segmentation models were trained for the preoperative (non-enhancing) tumor core, postoperative contrast-enhancing residual tumor, and resection cavity. Additionally, MR sequence classification and tumor type identification for contrast-enhancing lesions were explored using the DenseNet architecture. The models were integrated into a reporting pipeline, following the RANO 2.0 guidelines. Training was conducted on multicentric datasets comprising 2000 to 7000 patients, using a 5-fold cross-validation. Evaluation included patient-, voxel-, and object-wise metrics, with benchmarking against the latest BraTS challenge results. The segmentation models achieved average voxel-wise Dice scores of 87%, 66%, 70%, and 77% for the tumor core, non-enhancing tumor core, contrast-enhancing residual tumor, and resection cavity, respectively. Classification models reached 99.5% balanced accuracy in MR sequence classification and 80% in tumor type classification. The pipeline presented in this study enables robust, automated segmentation, MR sequence classification, and standardized report generation aligned with RANO 2.0 guidelines, enhancing postoperative evaluation and clinical decision-making. The proposed models and methods were integrated into Raidionics, open-source software platform for CNS tumor analysis, now including a dedicated module for postsurgical analysis.
A Stage-Aware Mixture of Experts Framework for Neurodegenerative Disease Progression Modelling
Aug 09, 2025Abstract:The long-term progression of neurodegenerative diseases is commonly conceptualized as a spatiotemporal diffusion process that consists of a graph diffusion process across the structural brain connectome and a localized reaction process within brain regions. However, modeling this progression remains challenging due to 1) the scarcity of longitudinal data obtained through irregular and infrequent subject visits and 2) the complex interplay of pathological mechanisms across brain regions and disease stages, where traditional models assume fixed mechanisms throughout disease progression. To address these limitations, we propose a novel stage-aware Mixture of Experts (MoE) framework that explicitly models how different contributing mechanisms dominate at different disease stages through time-dependent expert weighting.Data-wise, we utilize an iterative dual optimization method to properly estimate the temporal position of individual observations, constructing a co hort-level progression trajectory from irregular snapshots. Model-wise, we enhance the spatial component with an inhomogeneous graph neural diffusion model (IGND) that allows diffusivity to vary based on node states and time, providing more flexible representations of brain networks. We also introduce a localized neural reaction module to capture complex dynamics beyond standard processes.The resulting IGND-MoE model dynamically integrates these components across temporal states, offering a principled way to understand how stage-specific pathological mechanisms contribute to progression. The stage-wise weights yield novel clinical insights that align with literature, suggesting that graph-related processes are more influential at early stages, while other unknown physical processes become dominant later on.
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
P-Count: Persistence-based Counting of White Matter Hyperintensities in Brain MRI
Mar 20, 2024Abstract:White matter hyperintensities (WMH) are a hallmark of cerebrovascular disease and multiple sclerosis. Automated WMH segmentation methods enable quantitative analysis via estimation of total lesion load, spatial distribution of lesions, and number of lesions (i.e., number of connected components after thresholding), all of which are correlated with patient outcomes. While the two former measures can generally be estimated robustly, the number of lesions is highly sensitive to noise and segmentation mistakes -- even when small connected components are eroded or disregarded. In this article, we present P-Count, an algebraic WMH counting tool based on persistent homology that accounts for the topological features of WM lesions in a robust manner. Using computational geometry, P-Count takes the persistence of connected components into consideration, effectively filtering out the noisy WMH positives, resulting in a more accurate count of true lesions. We validated P-Count on the ISBI2015 longitudinal lesion segmentation dataset, where it produces significantly more accurate results than direct thresholding.
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI
Dec 08, 2023



Abstract:Brain atrophy and white matter hyperintensity (WMH) are critical neuroimaging features for ascertaining brain injury in cerebrovascular disease and multiple sclerosis. Automated segmentation and quantification is desirable but existing methods require high-resolution MRI with good signal-to-noise ratio (SNR). This precludes application to clinical and low-field portable MRI (pMRI) scans, thus hampering large-scale tracking of atrophy and WMH progression, especially in underserved areas where pMRI has huge potential. Here we present a method that segments white matter hyperintensity and 36 brain regions from scans of any resolution and contrast (including pMRI) without retraining. We show results on six public datasets and on a private dataset with paired high- and low-field scans (3T and 64mT), where we attain strong correlation between the WMH ($\rho$=.85) and hippocampal volumes (r=.89) estimated at both fields. Our method is publicly available as part of FreeSurfer, at: http://surfer.nmr.mgh.harvard.edu/fswiki/WMH-SynthSeg.
Segmentation of glioblastomas in early post-operative multi-modal MRI with deep neural networks
Apr 18, 2023Abstract:Extent of resection after surgery is one of the main prognostic factors for patients diagnosed with glioblastoma. To achieve this, accurate segmentation and classification of residual tumor from post-operative MR images is essential. The current standard method for estimating it is subject to high inter- and intra-rater variability, and an automated method for segmentation of residual tumor in early post-operative MRI could lead to a more accurate estimation of extent of resection. In this study, two state-of-the-art neural network architectures for pre-operative segmentation were trained for the task. The models were extensively validated on a multicenter dataset with nearly 1000 patients, from 12 hospitals in Europe and the United States. The best performance achieved was a 61\% Dice score, and the best classification performance was about 80\% balanced accuracy, with a demonstrated ability to generalize across hospitals. In addition, the segmentation performance of the best models was on par with human expert raters. The predicted segmentations can be used to accurately classify the patients into those with residual tumor, and those with gross total resection.
DeepBrainPrint: A Novel Contrastive Framework for Brain MRI Re-Identification
Feb 25, 2023



Abstract:Recent advances in MRI have led to the creation of large datasets. With the increase in data volume, it has become difficult to locate previous scans of the same patient within these datasets (a process known as re-identification). To address this issue, we propose an AI-powered medical imaging retrieval framework called DeepBrainPrint, which is designed to retrieve brain MRI scans of the same patient. Our framework is a semi-self-supervised contrastive deep learning approach with three main innovations. First, we use a combination of self-supervised and supervised paradigms to create an effective brain fingerprint from MRI scans that can be used for real-time image retrieval. Second, we use a special weighting function to guide the training and improve model convergence. Third, we introduce new imaging transformations to improve retrieval robustness in the presence of intensity variations (i.e. different scan contrasts), and to account for age and disease progression in patients. We tested DeepBrainPrint on a large dataset of T1-weighted brain MRIs from the Alzheimer's Disease Neuroimaging Initiative (ADNI) and on a synthetic dataset designed to evaluate retrieval performance with different image modalities. Our results show that DeepBrainPrint outperforms previous methods, including simple similarity metrics and more advanced contrastive deep learning frameworks.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022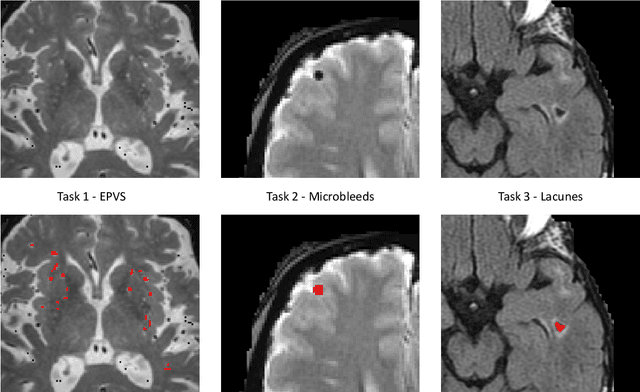
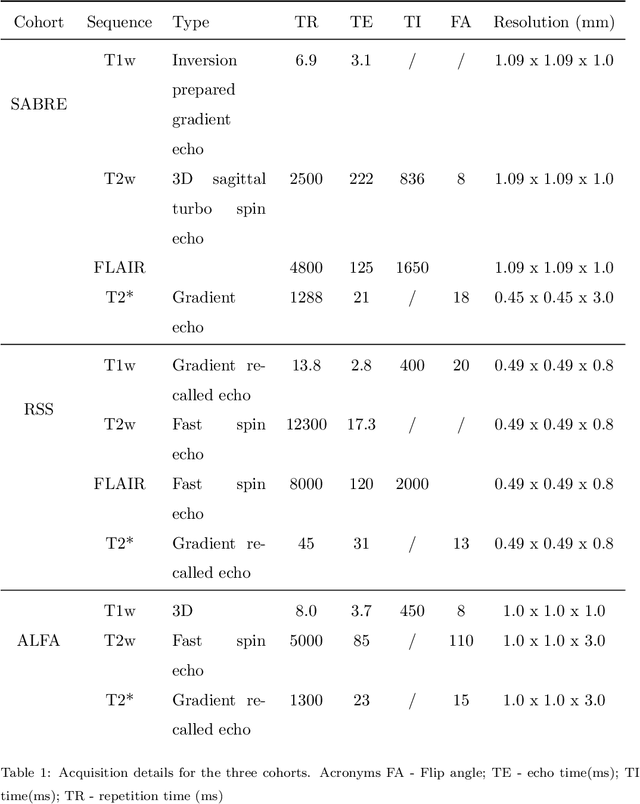
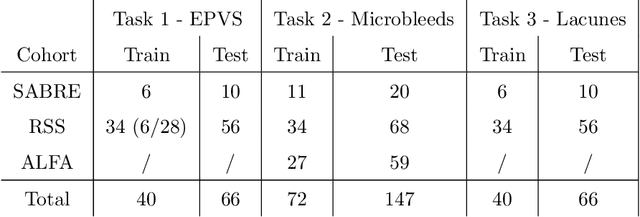
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Computer-aided diagnosis and prediction in brain disorders
Jun 29, 2022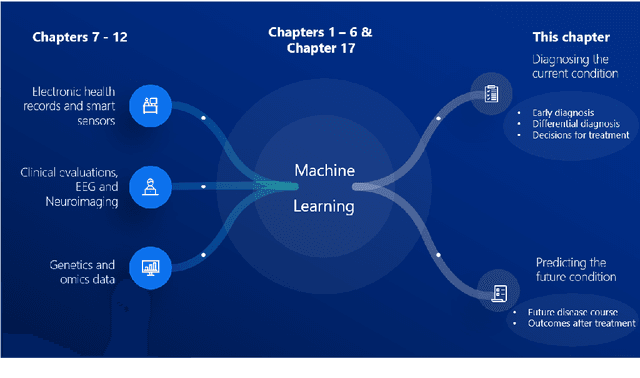
Abstract:Computer-aided methods have shown added value for diagnosing and predicting brain disorders and can thus support decision making in clinical care and treatment planning. This chapter will provide insight into the type of methods, their working, their input data - such as cognitive tests, imaging and genetic data - and the types of output they provide. We will focus on specific use cases for diagnosis, i.e. estimating the current 'condition' of the patient, such as early detection and diagnosis of dementia, differential diagnosis of brain tumours, and decision making in stroke. Regarding prediction, i.e. estimation of the future 'condition' of the patient, we will zoom in on use cases such as predicting the disease course in multiple sclerosis and predicting patient outcomes after treatment in brain cancer. Furthermore, based on these use cases, we will assess the current state-of-the-art methodology and highlight current efforts on benchmarking of these methods and the importance of open science therein. Finally, we assess the current clinical impact of computer-aided methods and discuss the required next steps to increase clinical impact.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge