Sébastien Ourselin
School of Biomedical Engineering and Imaging Sciences
Diffusion-Based Quality Control of Medical Image Segmentations across Organs
Nov 12, 2025



Abstract:Medical image segmentation using deep learning (DL) has enabled the development of automated analysis pipelines for large-scale population studies. However, state-of-the-art DL methods are prone to hallucinations, which can result in anatomically implausible segmentations. With manual correction impractical at scale, automated quality control (QC) techniques have to address the challenge. While promising, existing QC methods are organ-specific, limiting their generalizability and usability beyond their original intended task. To overcome this limitation, we propose no-new Quality Control (nnQC), a robust QC framework based on a diffusion-generative paradigm that self-adapts to any input organ dataset. Central to nnQC is a novel Team of Experts (ToE) architecture, where two specialized experts independently encode 3D spatial awareness, represented by the relative spatial position of an axial slice, and anatomical information derived from visual features from the original image. A weighted conditional module dynamically combines the pair of independent embeddings, or opinions to condition the sampling mechanism within a diffusion process, enabling the generation of a spatially aware pseudo-ground truth for predicting QC scores. Within its framework, nnQC integrates fingerprint adaptation to ensure adaptability across organs, datasets, and imaging modalities. We evaluated nnQC on seven organs using twelve publicly available datasets. Our results demonstrate that nnQC consistently outperforms state-of-the-art methods across all experiments, including cases where segmentation masks are highly degraded or completely missing, confirming its versatility and effectiveness across different organs.
Hydra: Marker-Free RGB-D Hand-Eye Calibration
Apr 29, 2025Abstract:This work presents an RGB-D imaging-based approach to marker-free hand-eye calibration using a novel implementation of the iterative closest point (ICP) algorithm with a robust point-to-plane (PTP) objective formulated on a Lie algebra. Its applicability is demonstrated through comprehensive experiments using three well known serial manipulators and two RGB-D cameras. With only three randomly chosen robot configurations, our approach achieves approximately 90% successful calibrations, demonstrating 2-3x higher convergence rates to the global optimum compared to both marker-based and marker-free baselines. We also report 2 orders of magnitude faster convergence time (0.8 +/- 0.4 s) for 9 robot configurations over other marker-free methods. Our method exhibits significantly improved accuracy (5 mm in task space) over classical approaches (7 mm in task space) whilst being marker-free. The benchmarking dataset and code are open sourced under Apache 2.0 License, and a ROS 2 integration with robot abstraction is provided to facilitate deployment.
Generative Medical Segmentation
Mar 27, 2024



Abstract:Rapid advancements in medical image segmentation performance have been significantly driven by the development of Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs). However, these models introduce high computational demands and often have limited ability to generalize across diverse medical imaging datasets. In this manuscript, we introduce Generative Medical Segmentation (GMS), a novel approach leveraging a generative model for image segmentation. Concretely, GMS employs a robust pre-trained Variational Autoencoder (VAE) to derive latent representations of both images and masks, followed by a mapping model that learns the transition from image to mask in the latent space. This process culminates in generating a precise segmentation mask within the image space using the pre-trained VAE decoder. The design of GMS leads to fewer learnable parameters in the model, resulting in a reduced computational burden and enhanced generalization capability. Our extensive experimental analysis across five public datasets in different medical imaging domains demonstrates GMS outperforms existing discriminative segmentation models and has remarkable domain generalization. Our experiments suggest GMS could set a new benchmark for medical image segmentation, offering a scalable and effective solution. GMS implementation and model weights are available at https://github.com/King-HAW/GMS.
UPL-SFDA: Uncertainty-aware Pseudo Label Guided Source-Free Domain Adaptation for Medical Image Segmentation
Sep 19, 2023



Abstract:Domain Adaptation (DA) is important for deep learning-based medical image segmentation models to deal with testing images from a new target domain. As the source-domain data are usually unavailable when a trained model is deployed at a new center, Source-Free Domain Adaptation (SFDA) is appealing for data and annotation-efficient adaptation to the target domain. However, existing SFDA methods have a limited performance due to lack of sufficient supervision with source-domain images unavailable and target-domain images unlabeled. We propose a novel Uncertainty-aware Pseudo Label guided (UPL) SFDA method for medical image segmentation. Specifically, we propose Target Domain Growing (TDG) to enhance the diversity of predictions in the target domain by duplicating the pre-trained model's prediction head multiple times with perturbations. The different predictions in these duplicated heads are used to obtain pseudo labels for unlabeled target-domain images and their uncertainty to identify reliable pseudo labels. We also propose a Twice Forward pass Supervision (TFS) strategy that uses reliable pseudo labels obtained in one forward pass to supervise predictions in the next forward pass. The adaptation is further regularized by a mean prediction-based entropy minimization term that encourages confident and consistent results in different prediction heads. UPL-SFDA was validated with a multi-site heart MRI segmentation dataset, a cross-modality fetal brain segmentation dataset, and a 3D fetal tissue segmentation dataset. It improved the average Dice by 5.54, 5.01 and 6.89 percentage points for the three tasks compared with the baseline, respectively, and outperformed several state-of-the-art SFDA methods.
A Dempster-Shafer approach to trustworthy AI with application to fetal brain MRI segmentation
Apr 05, 2022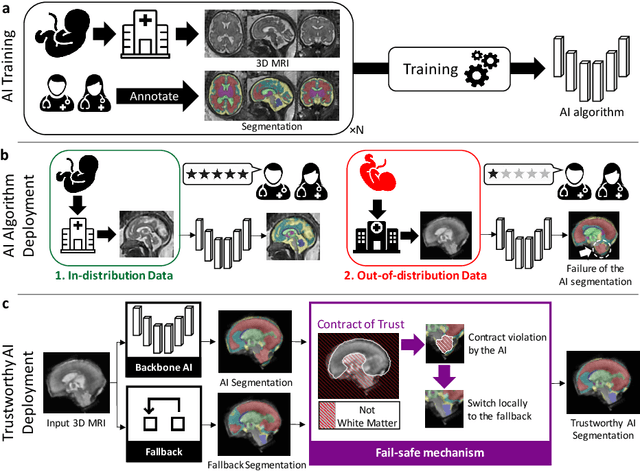
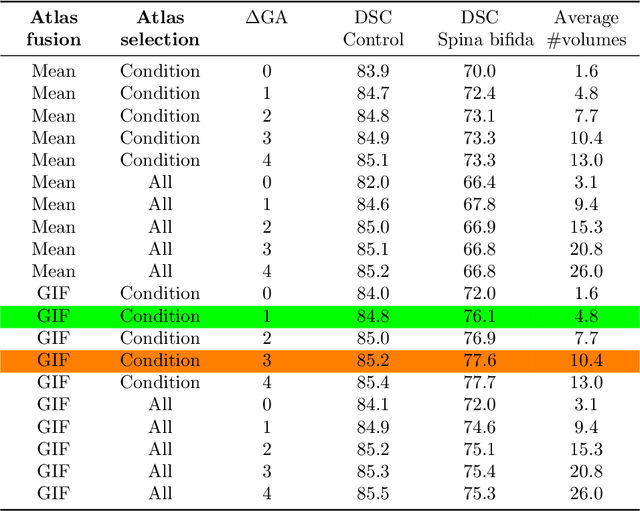
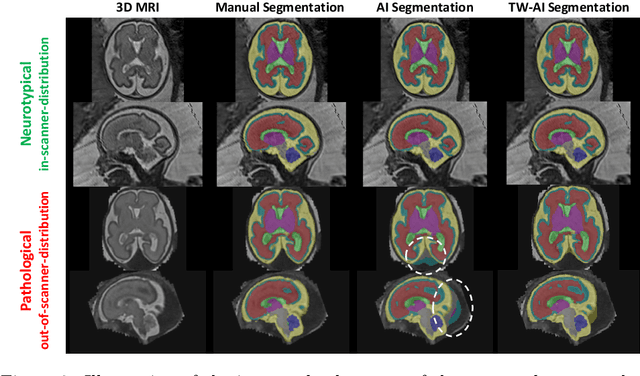
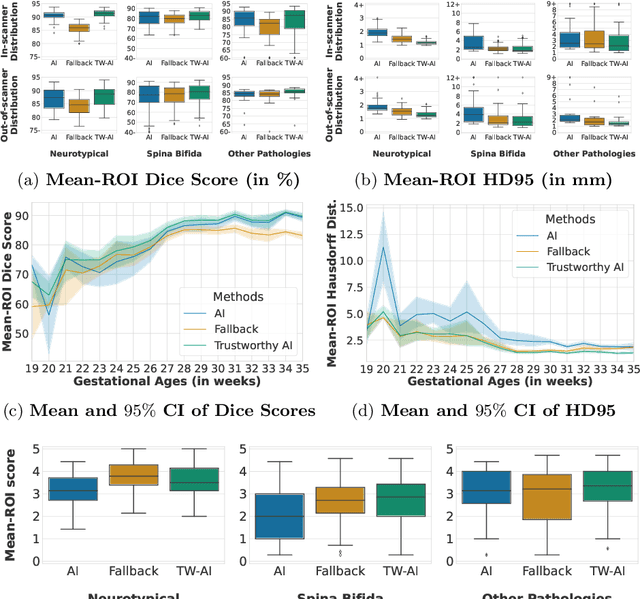
Abstract:Deep learning models for medical image segmentation can fail unexpectedly and spectacularly for pathological cases and for images acquired at different centers than those used for training, with labeling errors that violate expert knowledge about the anatomy and the intensity distribution of the regions to be segmented. Such errors undermine the trustworthiness of deep learning models developed for medical image segmentation. Mechanisms with a fallback method for detecting and correcting such failures are essential for safely translating this technology into clinics and are likely to be a requirement of future regulations on artificial intelligence (AI). Here, we propose a principled trustworthy AI theoretical framework and a practical system that can augment any backbone AI system using a fallback method and a fail-safe mechanism based on Dempster-Shafer theory. Our approach relies on an actionable definition of trustworthy AI. Our method automatically discards the voxel-level labeling predicted by the backbone AI that are likely to violate expert knowledge and relies on a fallback atlas-based segmentation method for those voxels. We demonstrate the effectiveness of the proposed trustworthy AI approach on the largest reported annotated dataset of fetal T2w MRI consisting of 540 manually annotated fetal brain 3D MRIs with neurotypical or abnormal brain development and acquired from 13 sources of data across 6 countries. We show that our trustworthy AI method improves the robustness of a state-of-the-art backbone AI for fetal brain MRI segmentation on MRIs acquired across various centers and for fetuses with various brain abnormalities.
Generalized Wasserstein Dice Loss, Test-time Augmentation, and Transformers for the BraTS 2021 challenge
Dec 24, 2021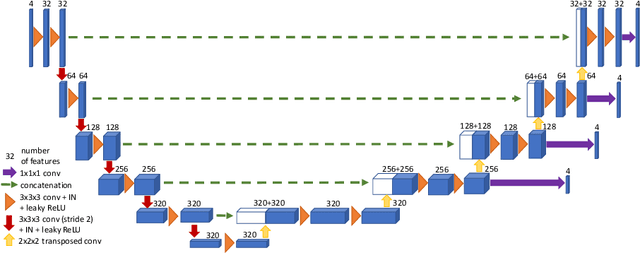
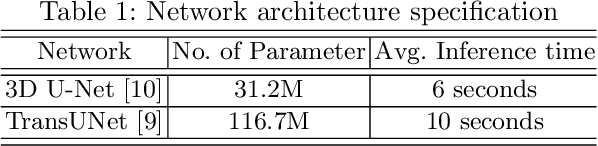
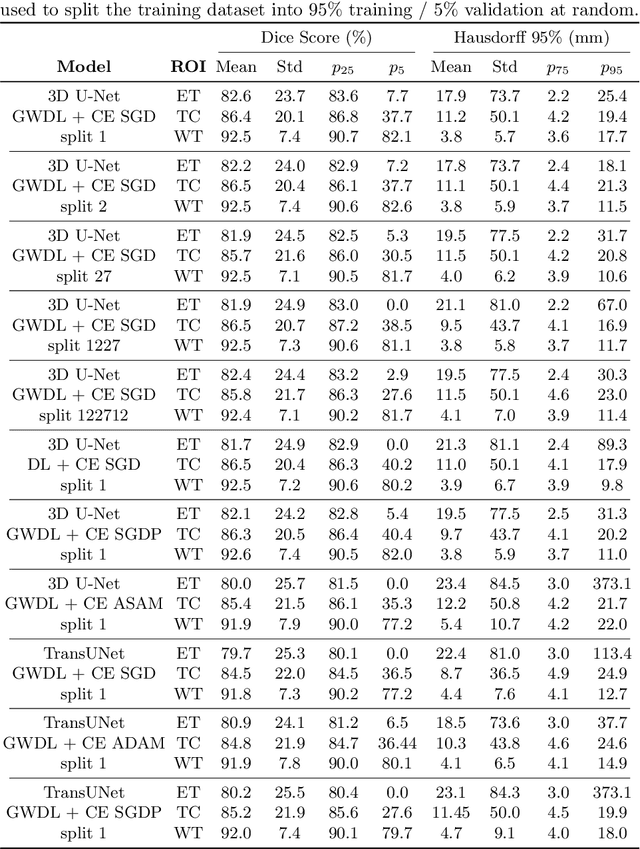
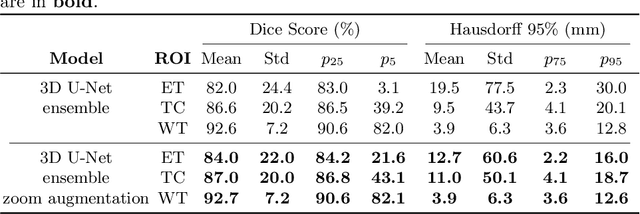
Abstract:Brain tumor segmentation from multiple Magnetic Resonance Imaging (MRI) modalities is a challenging task in medical image computation. The main challenges lie in the generalizability to a variety of scanners and imaging protocols. In this paper, we explore strategies to increase model robustness without increasing inference time. Towards this aim, we explore finding a robust ensemble from models trained using different losses, optimizers, and train-validation data split. Importantly, we explore the inclusion of a transformer in the bottleneck of the U-Net architecture. While we find transformer in the bottleneck performs slightly worse than the baseline U-Net in average, the generalized Wasserstein Dice loss consistently produces superior results. Further, we adopt an efficient test time augmentation strategy for faster and robust inference. Our final ensemble of seven 3D U-Nets with test-time augmentation produces an average dice score of 89.4% and an average Hausdorff 95% distance of 10.0 mm when evaluated on the BraTS 2021 testing dataset. Our code and trained models are publicly available at https://github.com/LucasFidon/TRABIT_BraTS2021.
Partial supervision for the FeTA challenge 2021
Nov 03, 2021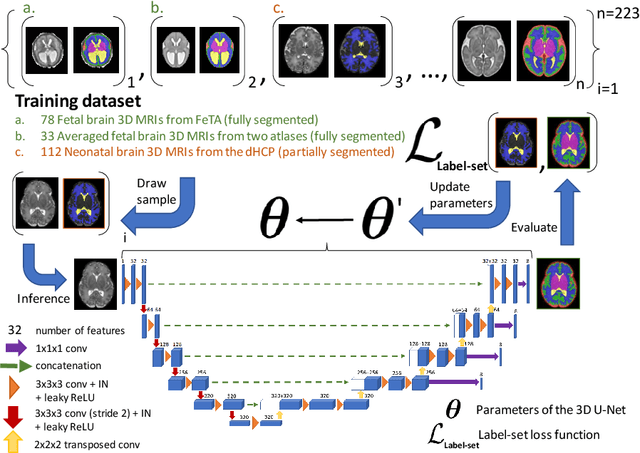
Abstract:This paper describes our method for our participation in the FeTA challenge2021 (team name: TRABIT). The performance of convolutional neural networks for medical image segmentation is thought to correlate positively with the number of training data. The FeTA challenge does not restrict participants to using only the provided training data but also allows for using other publicly available sources. Yet, open access fetal brain data remains limited. An advantageous strategy could thus be to expand the training data to cover broader perinatal brain imaging sources. Perinatal brain MRIs, other than the FeTA challenge data, that are currently publicly available, span normal and pathological fetal atlases as well as neonatal scans. However, perinatal brain MRIs segmented in different datasets typically come with different annotation protocols. This makes it challenging to combine those datasets to train a deep neural network. We recently proposed a family of loss functions, the label-set loss functions, for partially supervised learning. Label-set loss functions allow to train deep neural networks with partially segmented images, i.e. segmentations in which some classes may be grouped into super-classes. We propose to use label-set loss functions to improve the segmentation performance of a state-of-the-art deep learning pipeline for multi-class fetal brain segmentation by merging several publicly available datasets. To promote generalisability, our approach does not introduce any additional hyper-parameters tuning.
Homography-based Visual Servoing with Remote Center of Motion for Semi-autonomous Robotic Endoscope Manipulation
Oct 25, 2021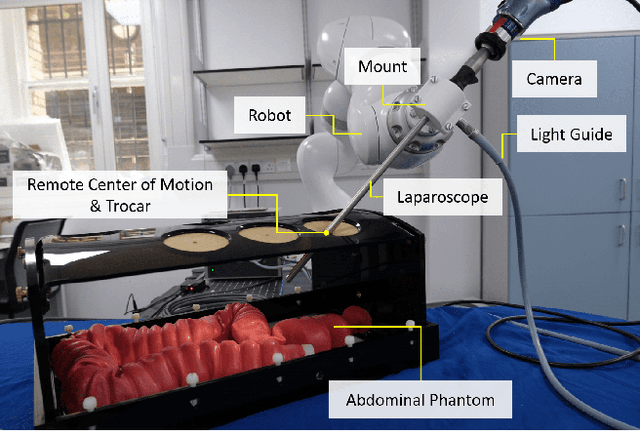
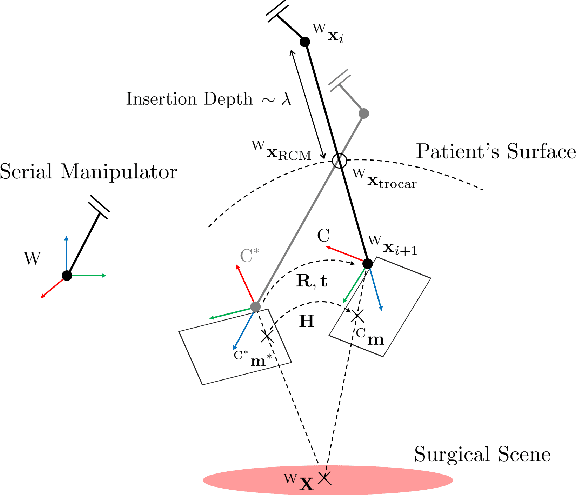
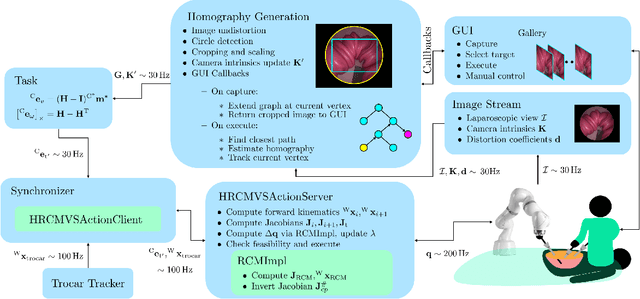
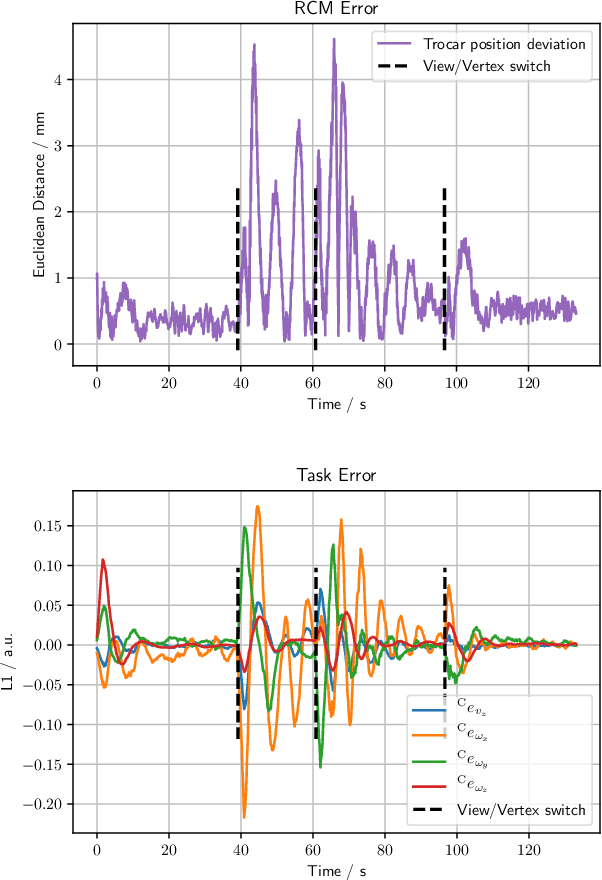
Abstract:The dominant visual servoing approaches in Minimally Invasive Surgery (MIS) follow single points or adapt the endoscope's field of view based on the surgical tools' distance. These methods rely on point positions with respect to the camera frame to infer a control policy. Deviating from the dominant methods, we formulate a robotic controller that allows for image-based visual servoing that requires neither explicit tool and camera positions nor any explicit image depth information. The proposed method relies on homography-based image registration, which changes the automation paradigm from point-centric towards surgical-scene-centric approach. It simultaneously respects a programmable Remote Center of Motion (RCM). Our approach allows a surgeon to build a graph of desired views, from which, once built, views can be manually selected and automatically servoed to irrespective of robot-patient frame transformation changes. We evaluate our method on an abdominal phantom and provide an open source ROS Moveit integration for use with any serial manipulator.
Deep Homography Estimation in Dynamic Surgical Scenes for Laparoscopic Camera Motion Extraction
Sep 30, 2021
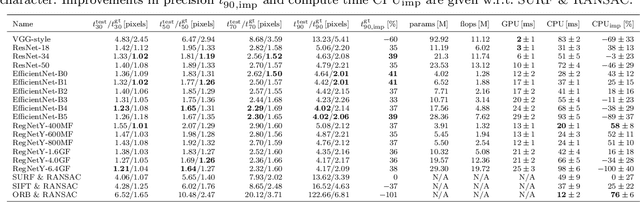
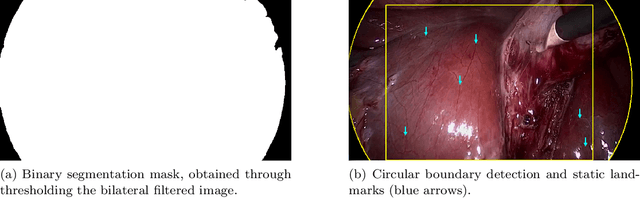
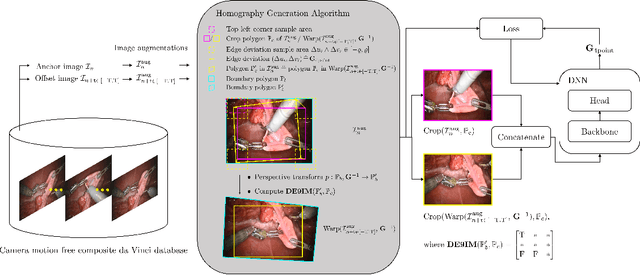
Abstract:Current laparoscopic camera motion automation relies on rule-based approaches or only focuses on surgical tools. Imitation Learning (IL) methods could alleviate these shortcomings, but have so far been applied to oversimplified setups. Instead of extracting actions from oversimplified setups, in this work we introduce a method that allows to extract a laparoscope holder's actions from videos of laparoscopic interventions. We synthetically add camera motion to a newly acquired dataset of camera motion free da Vinci surgery image sequences through the introduction of a novel homography generation algorithm. The synthetic camera motion serves as a supervisory signal for camera motion estimation that is invariant to object and tool motion. We perform an extensive evaluation of state-of-the-art (SOTA) Deep Neural Networks (DNNs) across multiple compute regimes, finding our method transfers from our camera motion free da Vinci surgery dataset to videos of laparoscopic interventions, outperforming classical homography estimation approaches in both, precision by 41%, and runtime on a CPU by 43%.
Distributionally Robust Segmentation of Abnormal Fetal Brain 3D MRI
Aug 09, 2021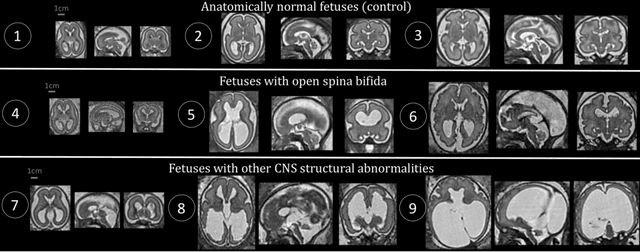
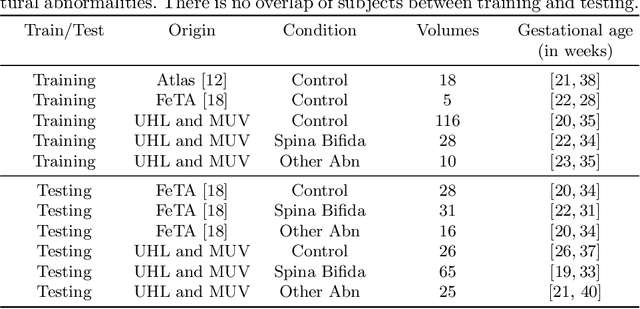
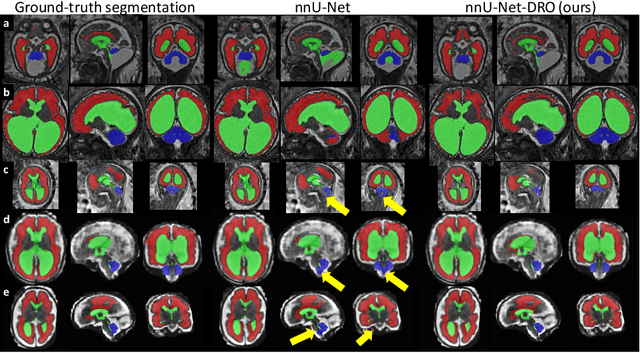
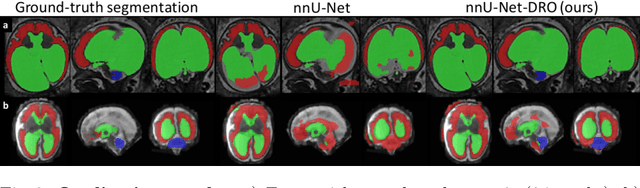
Abstract:The performance of deep neural networks typically increases with the number of training images. However, not all images have the same importance towards improved performance and robustness. In fetal brain MRI, abnormalities exacerbate the variability of the developing brain anatomy compared to non-pathological cases. A small number of abnormal cases, as is typically available in clinical datasets used for training, are unlikely to fairly represent the rich variability of abnormal developing brains. This leads machine learning systems trained by maximizing the average performance to be biased toward non-pathological cases. This problem was recently referred to as hidden stratification. To be suited for clinical use, automatic segmentation methods need to reliably achieve high-quality segmentation outcomes also for pathological cases. In this paper, we show that the state-of-the-art deep learning pipeline nnU-Net has difficulties to generalize to unseen abnormal cases. To mitigate this problem, we propose to train a deep neural network to minimize a percentile of the distribution of per-volume loss over the dataset. We show that this can be achieved by using Distributionally Robust Optimization (DRO). DRO automatically reweights the training samples with lower performance, encouraging nnU-Net to perform more consistently on all cases. We validated our approach using a dataset of 368 fetal brain T2w MRIs, including 124 MRIs of open spina bifida cases and 51 MRIs of cases with other severe abnormalities of brain development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge