Minjie Wang
Causal Discovery with Generalized Linear Models through Peeling Algorithms
Oct 25, 2023



Abstract:This article presents a novel method for causal discovery with generalized structural equation models suited for analyzing diverse types of outcomes, including discrete, continuous, and mixed data. Causal discovery often faces challenges due to unmeasured confounders that hinder the identification of causal relationships. The proposed approach addresses this issue by developing two peeling algorithms (bottom-up and top-down) to ascertain causal relationships and valid instruments. This approach first reconstructs a super-graph to represent ancestral relationships between variables, using a peeling algorithm based on nodewise GLM regressions that exploit relationships between primary and instrumental variables. Then, it estimates parent-child effects from the ancestral relationships using another peeling algorithm while deconfounding a child's model with information borrowed from its parents' models. The article offers a theoretical analysis of the proposed approach, which establishes conditions for model identifiability and provides statistical guarantees for accurately discovering parent-child relationships via the peeling algorithms. Furthermore, the article presents numerical experiments showcasing the effectiveness of our approach in comparison to state-of-the-art structure learning methods without confounders. Lastly, it demonstrates an application to Alzheimer's disease (AD), highlighting the utility of the method in constructing gene-to-gene and gene-to-disease regulatory networks involving Single Nucleotide Polymorphisms (SNPs) for healthy and AD subjects.
MuseGNN: Interpretable and Convergent Graph Neural Network Layers at Scale
Oct 19, 2023Abstract:Among the many variants of graph neural network (GNN) architectures capable of modeling data with cross-instance relations, an important subclass involves layers designed such that the forward pass iteratively reduces a graph-regularized energy function of interest. In this way, node embeddings produced at the output layer dually serve as both predictive features for solving downstream tasks (e.g., node classification) and energy function minimizers that inherit desirable inductive biases and interpretability. However, scaling GNN architectures constructed in this way remains challenging, in part because the convergence of the forward pass may involve models with considerable depth. To tackle this limitation, we propose a sampling-based energy function and scalable GNN layers that iteratively reduce it, guided by convergence guarantees in certain settings. We also instantiate a full GNN architecture based on these designs, and the model achieves competitive accuracy and scalability when applied to the largest publicly-available node classification benchmark exceeding 1TB in size.
ReFresh: Reducing Memory Access from Exploiting Stable Historical Embeddings for Graph Neural Network Training
Jan 19, 2023



Abstract:A key performance bottleneck when training graph neural network (GNN) models on large, real-world graphs is loading node features onto a GPU. Due to limited GPU memory, expensive data movement is necessary to facilitate the storage of these features on alternative devices with slower access (e.g. CPU memory). Moreover, the irregularity of graph structures contributes to poor data locality which further exacerbates the problem. Consequently, existing frameworks capable of efficiently training large GNN models usually incur a significant accuracy degradation because of the inevitable shortcuts involved. To address these limitations, we instead propose ReFresh, a general-purpose GNN mini-batch training framework that leverages a historical cache for storing and reusing GNN node embeddings instead of re-computing them through fetching raw features at every iteration. Critical to its success, the corresponding cache policy is designed, using a combination of gradient-based and staleness criteria, to selectively screen those embeddings which are relatively stable and can be cached, from those that need to be re-computed to reduce estimation errors and subsequent downstream accuracy loss. When paired with complementary system enhancements to support this selective historical cache, ReFresh is able to accelerate the training speed on large graph datasets such as ogbn-papers100M and MAG240M by 4.6x up to 23.6x and reduce the memory access by 64.5% (85.7% higher than a raw feature cache), with less than 1% influence on test accuracy.
DGI: Easy and Efficient Inference for GNNs
Nov 28, 2022Abstract:While many systems have been developed to train Graph Neural Networks (GNNs), efficient model inference and evaluation remain to be addressed. For instance, using the widely adopted node-wise approach, model evaluation can account for up to 94% of the time in the end-to-end training process due to neighbor explosion, which means that a node accesses its multi-hop neighbors. On the other hand, layer-wise inference avoids the neighbor explosion problem by conducting inference layer by layer such that the nodes only need their one-hop neighbors in each layer. However, implementing layer-wise inference requires substantial engineering efforts because users need to manually decompose a GNN model into layers for computation and split workload into batches to fit into device memory. In this paper, we develop Deep Graph Inference (DGI) -- a system for easy and efficient GNN model inference, which automatically translates the training code of a GNN model for layer-wise execution. DGI is general for various GNN models and different kinds of inference requests, and supports out-of-core execution on large graphs that cannot fit in CPU memory. Experimental results show that DGI consistently outperforms layer-wise inference across different datasets and hardware settings, and the speedup can be over 1,000x.
BiFeat: Supercharge GNN Training via Graph Feature Quantization
Jul 29, 2022



Abstract:Graph Neural Networks (GNNs) is a promising approach for applications with nonEuclidean data. However, training GNNs on large scale graphs with hundreds of millions nodes is both resource and time consuming. Different from DNNs, GNNs usually have larger memory footprints, and thus the GPU memory capacity and PCIe bandwidth are the main resource bottlenecks in GNN training. To address this problem, we present BiFeat: a graph feature quantization methodology to accelerate GNN training by significantly reducing the memory footprint and PCIe bandwidth requirement so that GNNs can take full advantage of GPU computing capabilities. Our key insight is that unlike DNN, GNN is less prone to the information loss of input features caused by quantization. We identify the main accuracy impact factors in graph feature quantization and theoretically prove that BiFeat training converges to a network where the loss is within $\epsilon$ of the optimal loss of uncompressed network. We perform extensive evaluation of BiFeat using several popular GNN models and datasets, including GraphSAGE on MAG240M, the largest public graph dataset. The results demonstrate that BiFeat achieves a compression ratio of more than 30 and improves GNN training speed by 200%-320% with marginal accuracy loss. In particular, BiFeat achieves a record by training GraphSAGE on MAG240M within one hour using only four GPUs.
CEP3: Community Event Prediction with Neural Point Process on Graph
May 21, 2022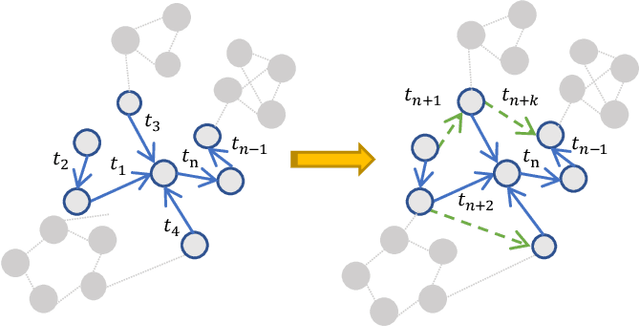
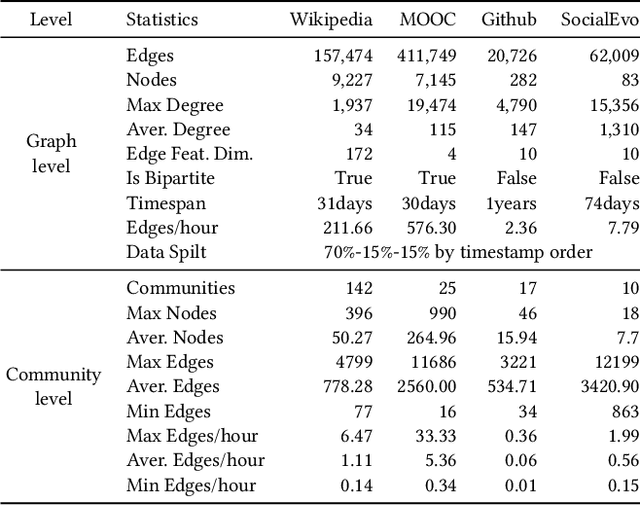
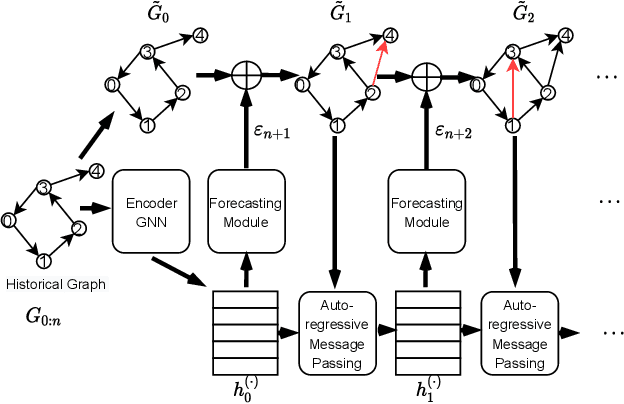
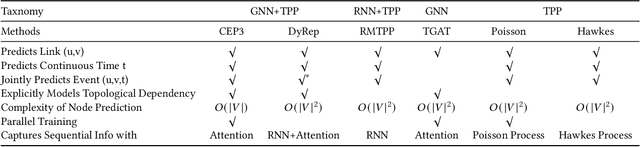
Abstract:Many real world applications can be formulated as event forecasting on Continuous Time Dynamic Graphs (CTDGs) where the occurrence of a timed event between two entities is represented as an edge along with its occurrence timestamp in the graphs.However, most previous works approach the problem in compromised settings, either formulating it as a link prediction task on the graph given the event time or a time prediction problem given which event will happen next. In this paper, we propose a novel model combining Graph Neural Networks and Marked Temporal Point Process (MTPP) that jointly forecasts multiple link events and their timestamps on communities over a CTDG. Moreover, to scale our model to large graphs, we factorize the jointly event prediction problem into three easier conditional probability modeling problems.To evaluate the effectiveness of our model and the rationale behind such a decomposition, we establish a set of benchmarks and evaluation metrics for this event forecasting task. Our experiments demonstrate the superior performance of our model in terms of both model accuracy and training efficiency.
Gaussian Graphical Model Selection for Huge Data via Minipatch Learning
Oct 22, 2021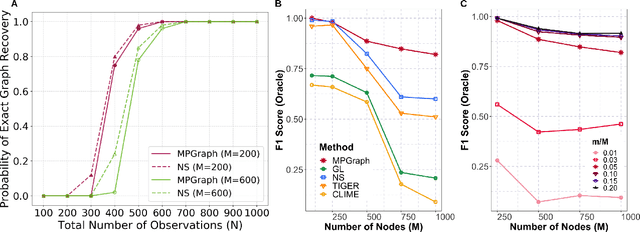
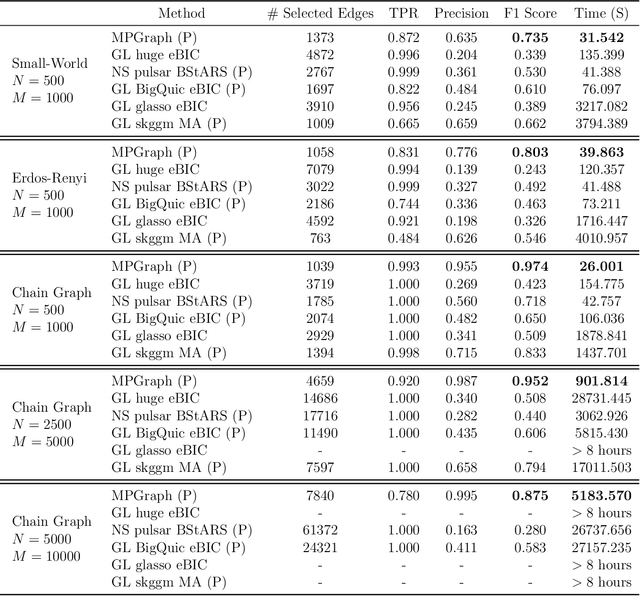
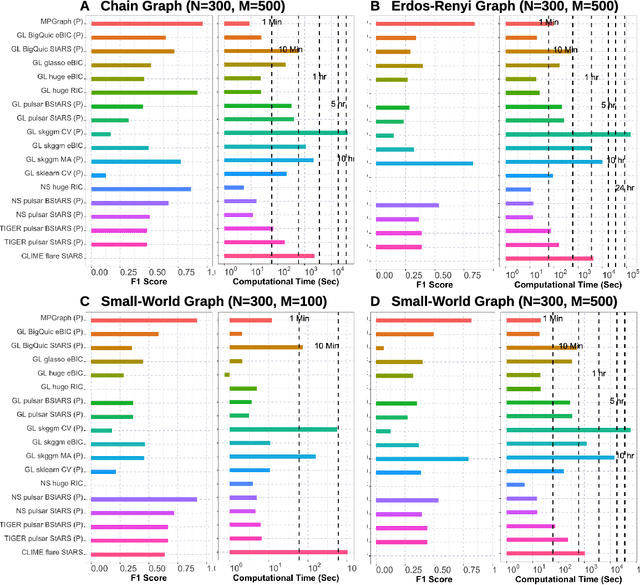
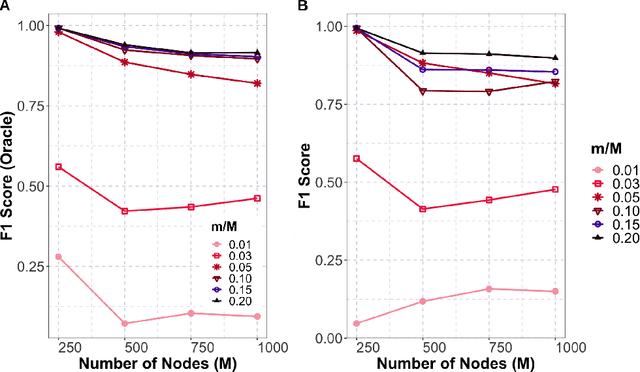
Abstract:Gaussian graphical models are essential unsupervised learning techniques to estimate conditional dependence relationships between sets of nodes. While graphical model selection is a well-studied problem with many popular techniques, there are typically three key practical challenges: i) many existing methods become computationally intractable in huge-data settings with tens of thousands of nodes; ii) the need for separate data-driven tuning hyperparameter selection procedures considerably adds to the computational burden; iii) the statistical accuracy of selected edges often deteriorates as the dimension and/or the complexity of the underlying graph structures increase. We tackle these problems by proposing the Minipatch Graph (MPGraph) estimator. Our approach builds upon insights from the latent variable graphical model problem and utilizes ensembles of thresholded graph estimators fit to tiny, random subsets of both the observations and the nodes, termed minipatches. As estimates are fit on small problems, our approach is computationally fast with integrated stability-based hyperparameter tuning. Additionally, we prove that under certain conditions our MPGraph algorithm achieves finite-sample graph selection consistency. We compare our approach to state-of-the-art computational approaches to Gaussian graphical model selection including the BigQUIC algorithm, and empirically demonstrate that our approach is not only more accurate but also extensively faster for huge graph selection problems.
Thresholded Graphical Lasso Adjusts for Latent Variables: Application to Functional Neural Connectivity
Apr 13, 2021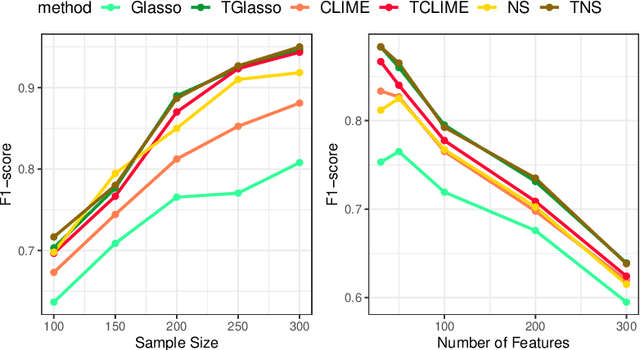

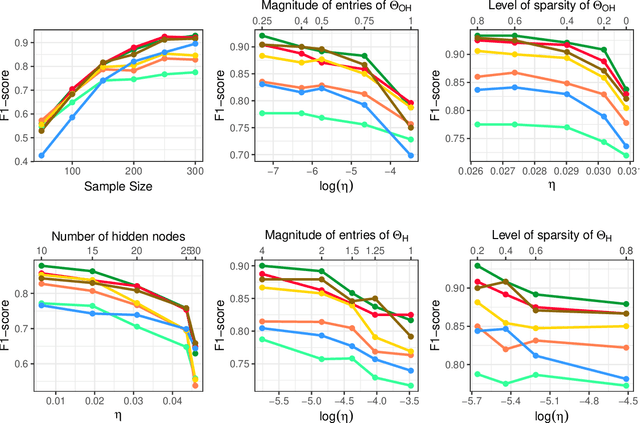
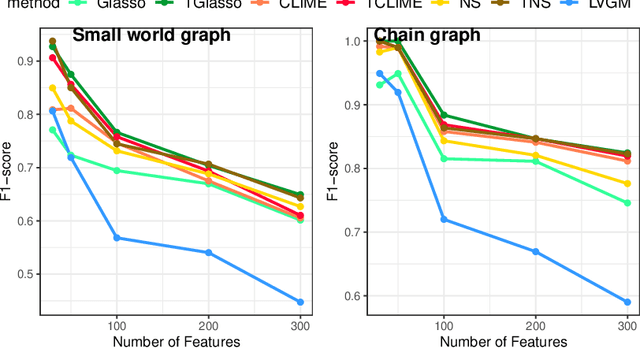
Abstract:In neuroscience, researchers seek to uncover the connectivity of neurons from large-scale neural recordings or imaging; often people employ graphical model selection and estimation techniques for this purpose. But, existing technologies can only record from a small subset of neurons leading to a challenging problem of graph selection in the presence of extensive latent variables. Chandrasekaran et al. (2012) proposed a convex program to address this problem that poses challenges from both a computational and statistical perspective. To solve this problem, we propose an incredibly simple solution: apply a hard thresholding operator to existing graph selection methods. Conceptually simple and computationally attractive, we demonstrate that thresholding the graphical Lasso, neighborhood selection, or CLIME estimators have superior theoretical properties in terms of graph selection consistency as well as stronger empirical results than existing approaches for the latent variable graphical model problem. We also demonstrate the applicability of our approach through a neuroscience case study on calcium-imaging data to estimate functional neural connections.
DistDGL: Distributed Graph Neural Network Training for Billion-Scale Graphs
Oct 16, 2020
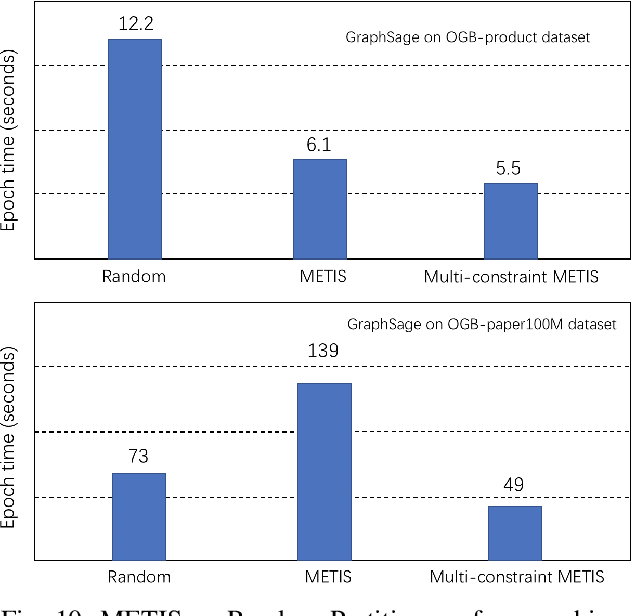
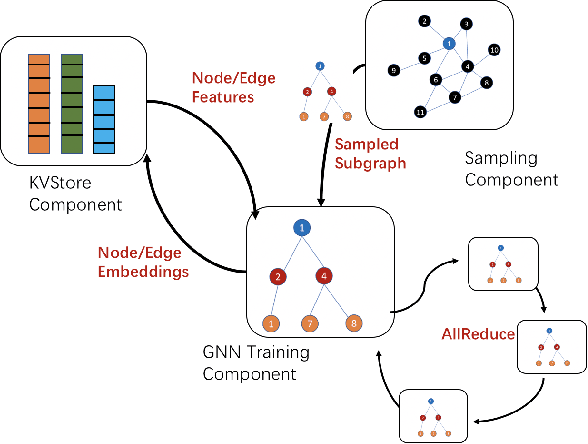
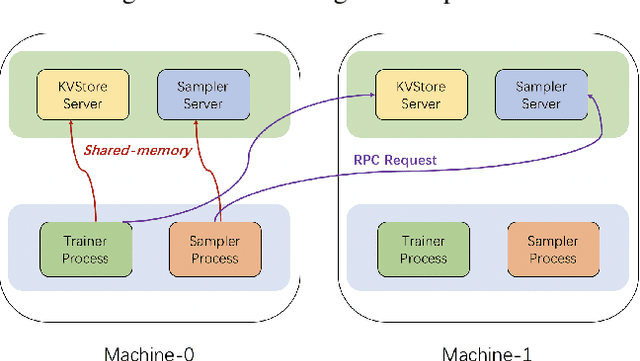
Abstract:Graph neural networks (GNN) have shown great success in learning from graph-structured data. They are widely used in various applications, such as recommendation, fraud detection, and search. In these domains, the graphs are typically large, containing hundreds of millions of nodes and several billions of edges. To tackle this challenge, we develop DistDGL, a system for training GNNs in a mini-batch fashion on a cluster of machines. DistDGL is based on the Deep Graph Library (DGL), a popular GNN development framework. DistDGL distributes the graph and its associated data (initial features and embeddings) across the machines and uses this distribution to derive a computational decomposition by following an owner-compute rule. DistDGL follows a synchronous training approach and allows ego-networks forming the mini-batches to include non-local nodes. To minimize the overheads associated with distributed computations, DistDGL uses a high-quality and light-weight min-cut graph partitioning algorithm along with multiple balancing constraints. This allows it to reduce communication overheads and statically balance the computations. It further reduces the communication by replicating halo nodes and by using sparse embedding updates. The combination of these design choices allows DistDGL to train high-quality models while achieving high parallel efficiency and memory scalability. We demonstrate our optimizations on both inductive and transductive GNN models. Our results show that DistDGL achieves linear speedup without compromising model accuracy and requires only 13 seconds to complete a training epoch for a graph with 100 million nodes and 3 billion edges on a cluster with 16 machines.
FeatGraph: A Flexible and Efficient Backend for Graph Neural Network Systems
Sep 29, 2020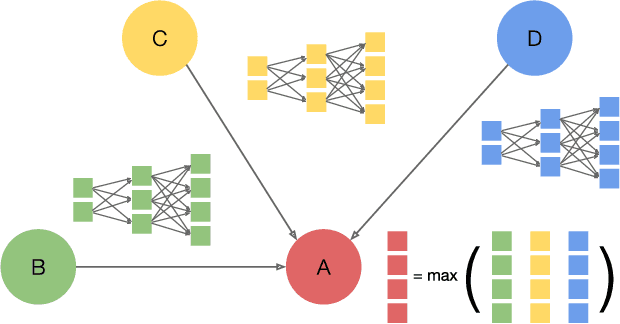
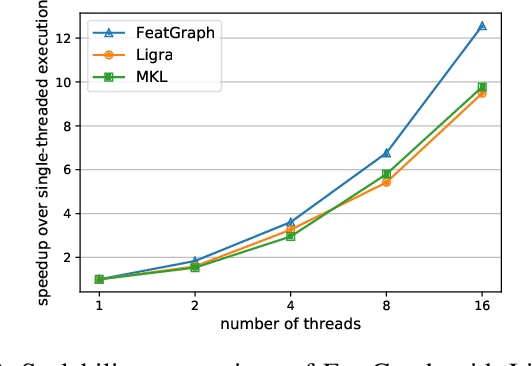
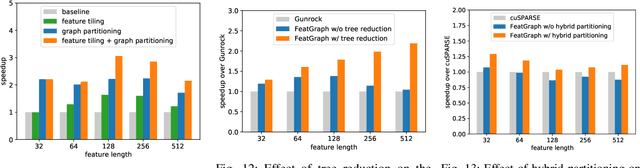
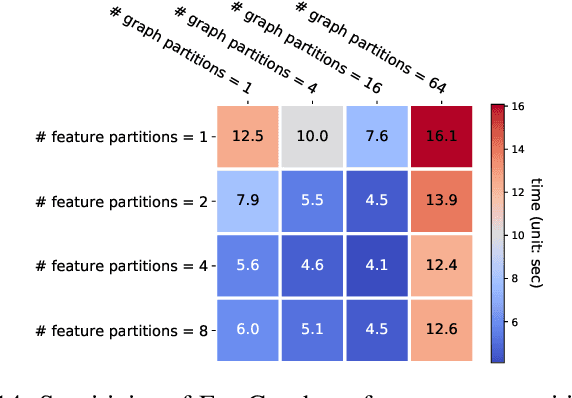
Abstract:Graph neural networks (GNNs) are gaining increasing popularity as a promising approach to machine learning on graphs. Unlike traditional graph workloads where each vertex/edge is associated with a scalar, GNNs attach a feature tensor to each vertex/edge. This additional feature dimension, along with consequently more complex vertex- and edge-wise computations, has enormous implications on locality and parallelism, which existing graph processing systems fail to exploit. This paper proposes FeatGraph to accelerate GNN workloads by co-optimizing graph traversal and feature dimension computation. FeatGraph provides a flexible programming interface to express diverse GNN models by composing coarse-grained sparse templates with fine-grained user-defined functions (UDFs) on each vertex/edge. FeatGraph incorporates optimizations for graph traversal into the sparse templates and allows users to specify optimizations for UDFs with a feature dimension schedule (FDS). FeatGraph speeds up end-to-end GNN training and inference by up to 32x on CPU and 7x on GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge