Mei Han
Accurate and Robust Lesion RECIST Diameter Prediction and Segmentation with Transformers
Aug 28, 2022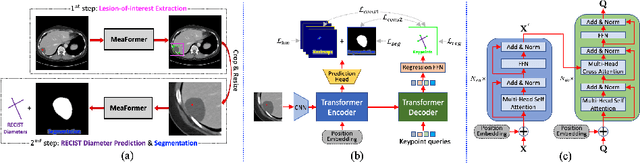
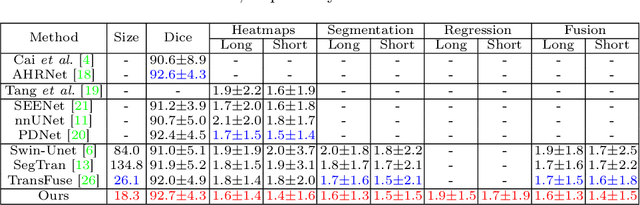
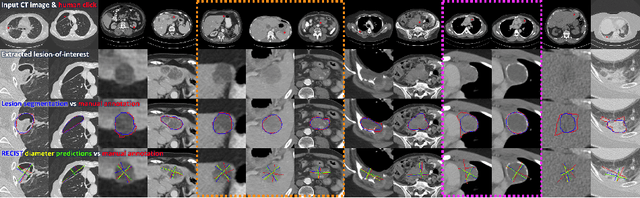
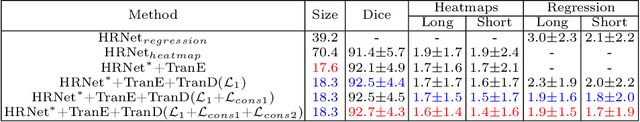
Abstract:Automatically measuring lesion/tumor size with RECIST (Response Evaluation Criteria In Solid Tumors) diameters and segmentation is important for computer-aided diagnosis. Although it has been studied in recent years, there is still space to improve its accuracy and robustness, such as (1) enhancing features by incorporating rich contextual information while keeping a high spatial resolution and (2) involving new tasks and losses for joint optimization. To reach this goal, this paper proposes a transformer-based network (MeaFormer, Measurement transFormer) for lesion RECIST diameter prediction and segmentation (LRDPS). It is formulated as three correlative and complementary tasks: lesion segmentation, heatmap prediction, and keypoint regression. To the best of our knowledge, it is the first time to use keypoint regression for RECIST diameter prediction. MeaFormer can enhance high-resolution features by employing transformers to capture their long-range dependencies. Two consistency losses are introduced to explicitly build relationships among these tasks for better optimization. Experiments show that MeaFormer achieves the state-of-the-art performance of LRDPS on the large-scale DeepLesion dataset and produces promising results of two downstream clinic-relevant tasks, i.e., 3D lesion segmentation and RECIST assessment in longitudinal studies.
PieTrack: An MOT solution based on synthetic data training and self-supervised domain adaptation
Jul 22, 2022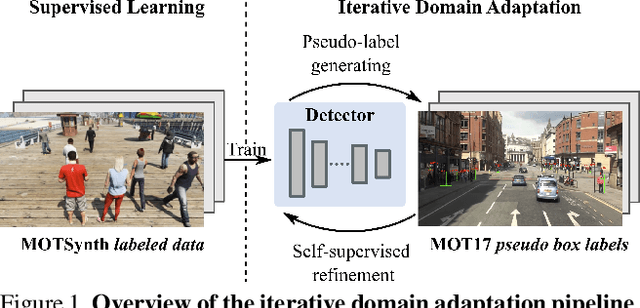
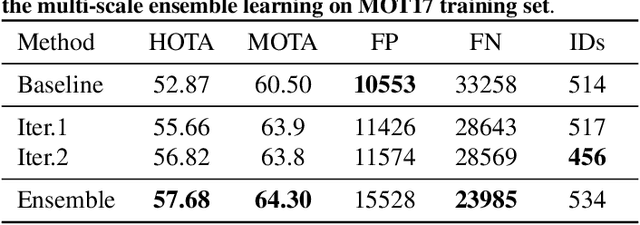
Abstract:In order to cope with the increasing demand for labeling data and privacy issues with human detection, synthetic data has been used as a substitute and showing promising results in human detection and tracking tasks. We participate in the 7th Workshop on Benchmarking Multi-Target Tracking (BMTT), themed on "How Far Can Synthetic Data Take us"? Our solution, PieTrack, is developed based on synthetic data without using any pre-trained weights. We propose a self-supervised domain adaptation method that enables mitigating the domain shift issue between the synthetic (e.g., MOTSynth) and real data (e.g., MOT17) without involving extra human labels. By leveraging the proposed multi-scale ensemble inference, we achieved a final HOTA score of 58.7 on the MOT17 testing set, ranked third place in the challenge.
Localized Adversarial Domain Generalization
May 09, 2022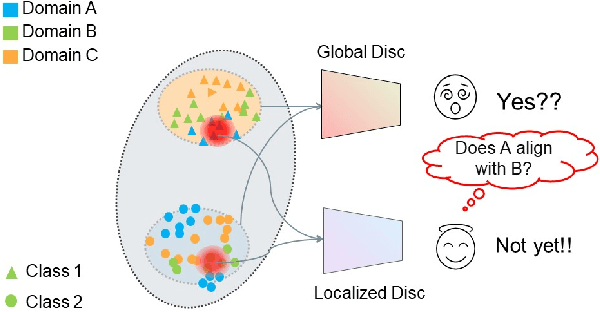
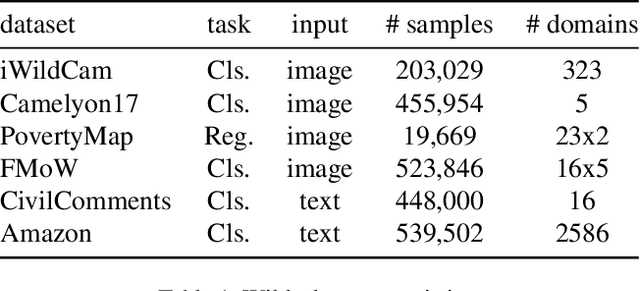
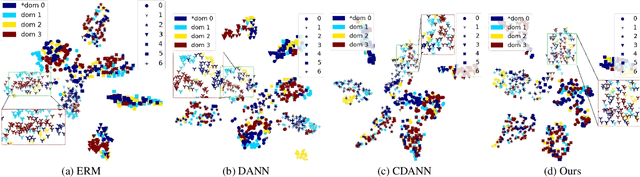
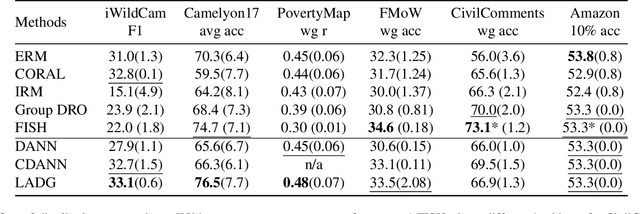
Abstract:Deep learning methods can struggle to handle domain shifts not seen in training data, which can cause them to not generalize well to unseen domains. This has led to research attention on domain generalization (DG), which aims to the model's generalization ability to out-of-distribution. Adversarial domain generalization is a popular approach to DG, but conventional approaches (1) struggle to sufficiently align features so that local neighborhoods are mixed across domains; and (2) can suffer from feature space over collapse which can threaten generalization performance. To address these limitations, we propose localized adversarial domain generalization with space compactness maintenance~(LADG) which constitutes two major contributions. First, we propose an adversarial localized classifier as the domain discriminator, along with a principled primary branch. This constructs a min-max game whereby the aim of the featurizer is to produce locally mixed domains. Second, we propose to use a coding-rate loss to alleviate feature space over collapse. We conduct comprehensive experiments on the Wilds DG benchmark to validate our approach, where LADG outperforms leading competitors on most datasets.
Scalable Semi-supervised Landmark Localization for X-ray Images using Few-shot Deep Adaptive Graph
Apr 29, 2021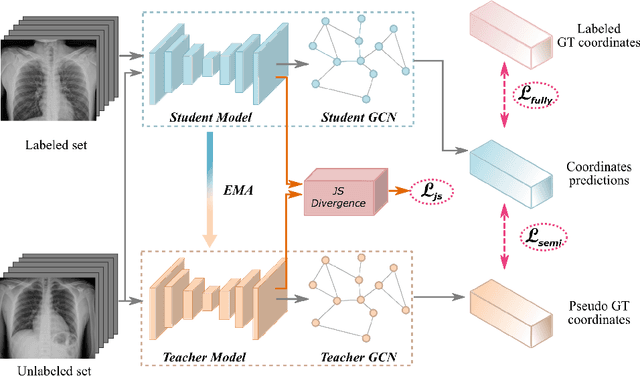
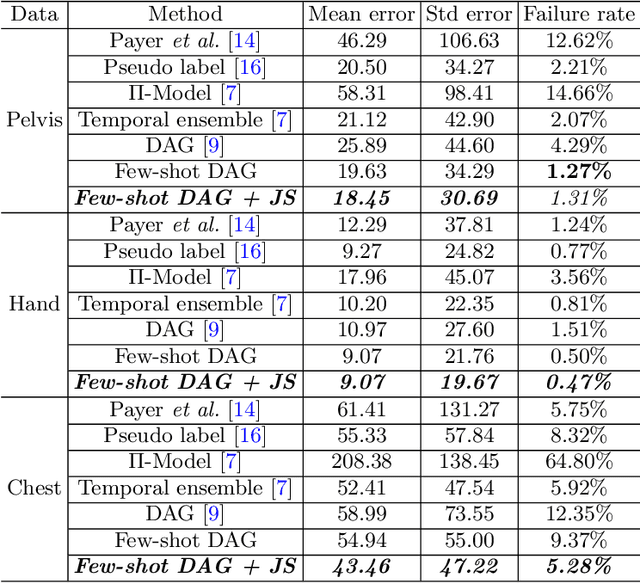
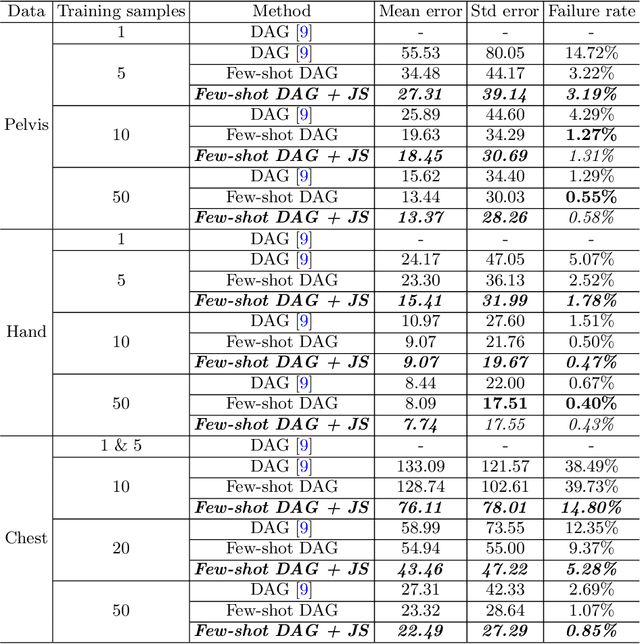
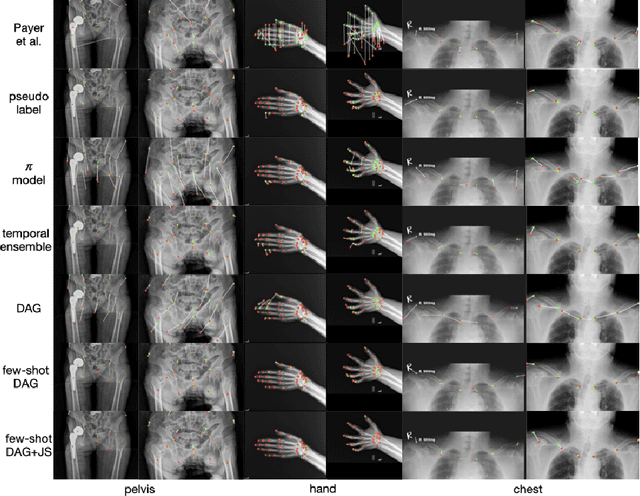
Abstract:Landmark localization plays an important role in medical image analysis. Learning based methods, including CNN and GCN, have demonstrated the state-of-the-art performance. However, most of these methods are fully-supervised and heavily rely on manual labeling of a large training dataset. In this paper, based on a fully-supervised graph-based method, DAG, we proposed a semi-supervised extension of it, termed few-shot DAG, \ie five-shot DAG. It first trains a DAG model on the labeled data and then fine-tunes the pre-trained model on the unlabeled data with a teacher-student SSL mechanism. In addition to the semi-supervised loss, we propose another loss using JS divergence to regulate the consistency of the intermediate feature maps. We extensively evaluated our method on pelvis, hand and chest landmark detection tasks. Our experiment results demonstrate consistent and significant improvements over previous methods.
Deep Implicit Statistical Shape Models for 3D Medical Image Delineation
Apr 07, 2021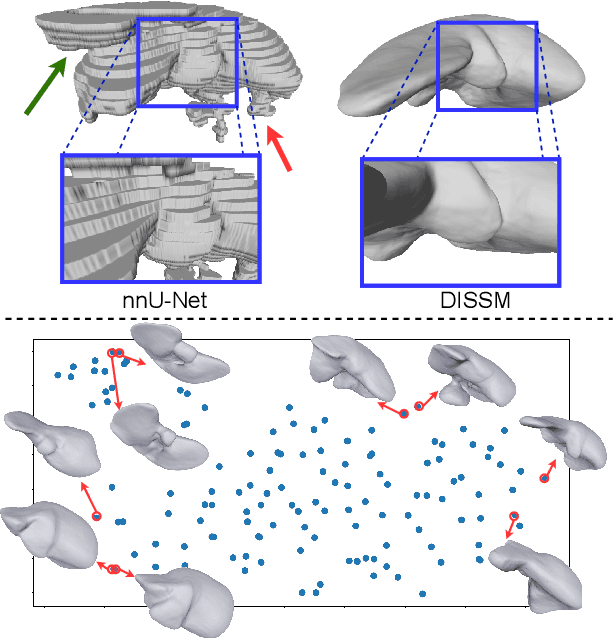
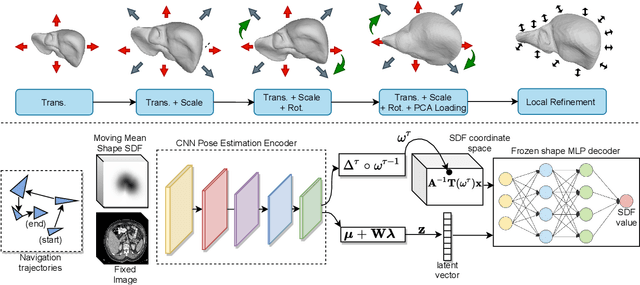
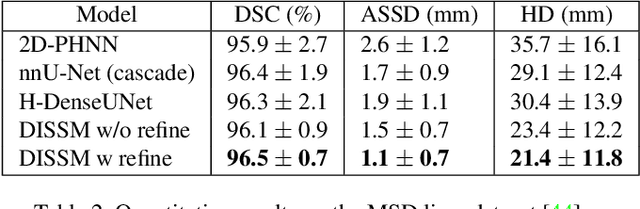
Abstract:3D delineation of anatomical structures is a cardinal goal in medical imaging analysis. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Today fully-convolutional networks (FCNs), while dominant, do not offer these capabilities. We present deep implicit statistical shape models (DISSMs), a new approach to delineation that marries the representation power of convolutional neural networks (CNNs) with the robustness of SSMs. DISSMs use a deep implicit surface representation to produce a compact and descriptive shape latent space that permits statistical models of anatomical variance. To reliably fit anatomically plausible shapes to an image, we introduce a novel rigid and non-rigid pose estimation pipeline that is modelled as a Markov decision process(MDP). We outline a training regime that includes inverted episodic training and a deep realization of marginal space learning (MSL). Intra-dataset experiments on the task of pathological liver segmentation demonstrate that DISSMs can perform more robustly than three leading FCN models, including nnU-Net: reducing the mean Hausdorff distance (HD) by 7.7-14.3mm and improving the worst case Dice-Sorensen coefficient (DSC) by 1.2-2.3%. More critically, cross-dataset experiments on a dataset directly reflecting clinical deployment scenarios demonstrate that DISSMs improve the mean DSC and HD by 3.5-5.9% and 12.3-24.5mm, respectively, and the worst-case DSC by 5.4-7.3%. These improvements are over and above any benefits from representing delineations with high-quality surface.
Hetero-Modal Learning and Expansive Consistency Constraints for Semi-Supervised Detection from Multi-Sequence Data
Mar 24, 2021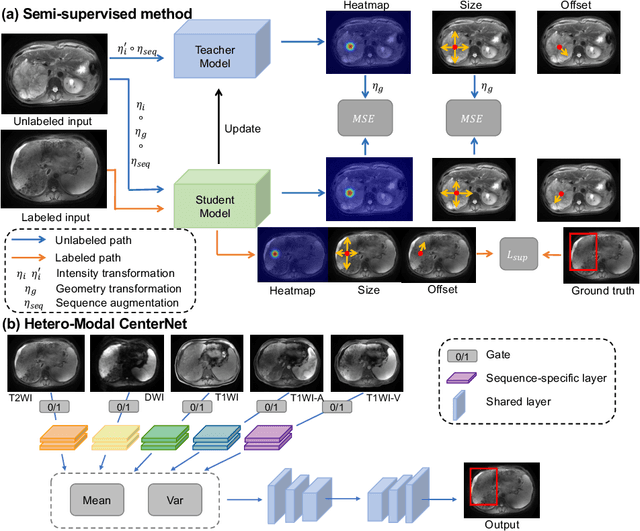
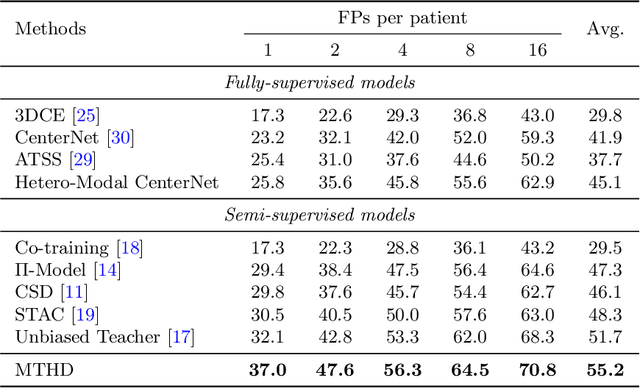
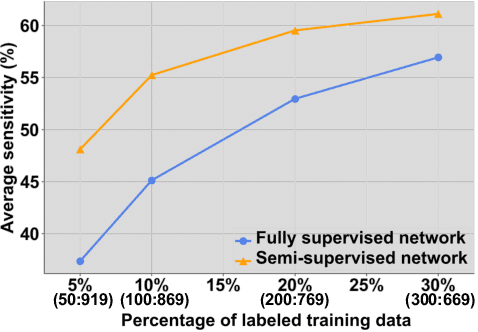
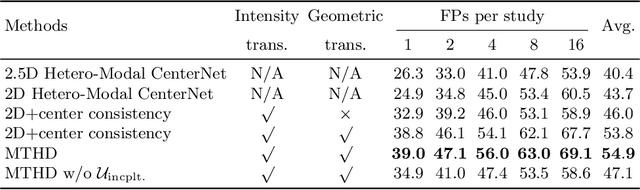
Abstract:Lesion detection serves a critical role in early diagnosis and has been well explored in recent years due to methodological advancesand increased data availability. However, the high costs of annotations hinder the collection of large and completely labeled datasets, motivating semi-supervised detection approaches. In this paper, we introduce mean teacher hetero-modal detection (MTHD), which addresses two important gaps in current semi-supervised detection. First, it is not obvious how to enforce unlabeled consistency constraints across the very different outputs of various detectors, which has resulted in various compromises being used in the state of the art. Using an anchor-free framework, MTHD formulates a mean teacher approach without such compromises, enforcing consistency on the soft-output of object centers and size. Second, multi-sequence data is often critical, e.g., for abdominal lesion detection, but unlabeled data is often missing sequences. To deal with this, MTHD incorporates hetero-modal learning in its framework. Unlike prior art, MTHD is able to incorporate an expansive set of consistency constraints that include geometric transforms and random sequence combinations. We train and evaluate MTHD on liver lesion detection using the largest MR lesion dataset to date (1099 patients with >5000 volumes). MTHD surpasses the best fully-supervised and semi-supervised competitors by 10.1% and 3.5%, respectively, in average sensitivity.
SegAttnGAN: Text to Image Generation with Segmentation Attention
May 25, 2020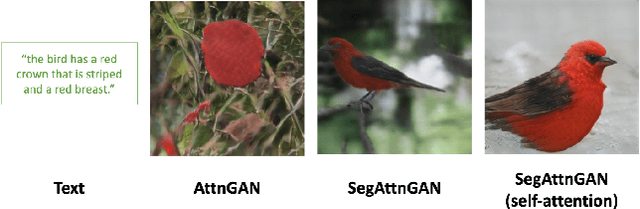
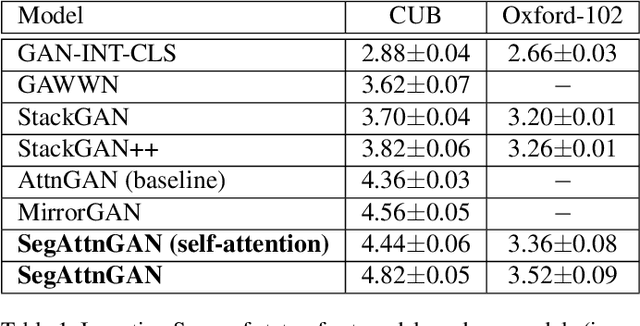
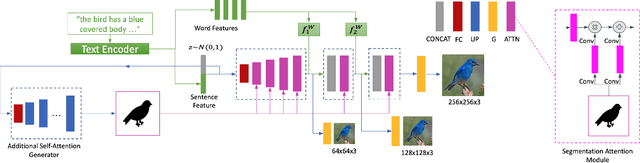

Abstract:In this paper, we propose a novel generative network (SegAttnGAN) that utilizes additional segmentation information for the text-to-image synthesis task. As the segmentation data introduced to the model provides useful guidance on the generator training, the proposed model can generate images with better realism quality and higher quantitative measures compared with the previous state-of-art methods. We achieved Inception Score of 4.84 on the CUB dataset and 3.52 on the Oxford-102 dataset. Besides, we tested the self-attention SegAttnGAN which uses generated segmentation data instead of masks from datasets for attention and achieved similar high-quality results, suggesting that our model can be adapted for the text-to-image synthesis task.
Handwritten Chinese Font Generation with Collaborative Stroke Refinement
May 06, 2019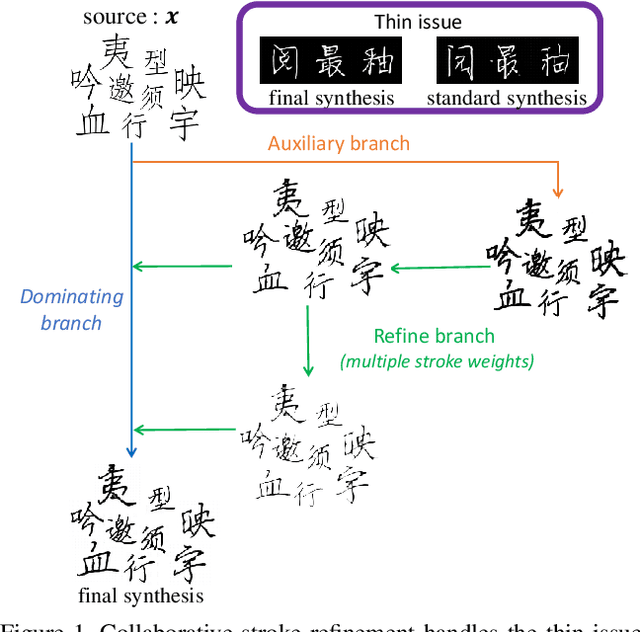
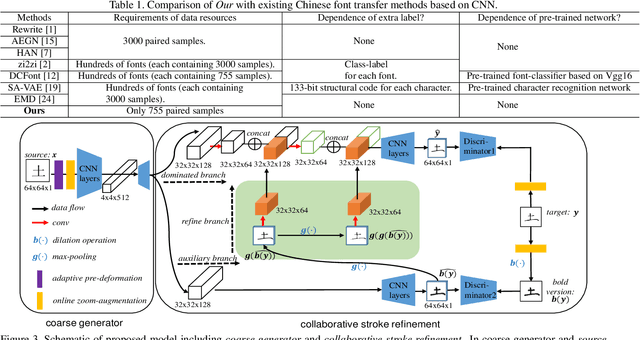
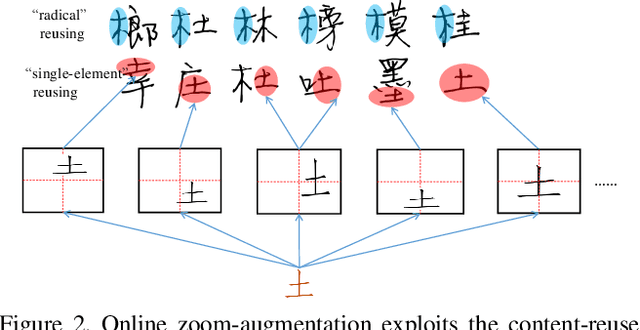

Abstract:Automatic character generation is an appealing solution for new typeface design, especially for Chinese typefaces including over 3700 most commonly-used characters. This task has two main pain points: (i) handwritten characters are usually associated with thin strokes of few information and complex structure which are error prone during deformation; (ii) thousands of characters with various shapes are needed to synthesize based on a few manually designed characters. To solve those issues, we propose a novel convolutional-neural-network-based model with three main techniques: collaborative stroke refinement, using collaborative training strategy to recover the missing or broken strokes; online zoom-augmentation, taking the advantage of the content-reuse phenomenon to reduce the size of training set; and adaptive pre-deformation, standardizing and aligning the characters. The proposed model needs only 750 paired training samples; no pre-trained network, extra dataset resource or labels is needed. Experimental results show that the proposed method significantly outperforms the state-of-the-art methods under the practical restriction on handwritten font synthesis.
Prior-aware Neural Network for Partially-Supervised Multi-Organ Segmentation
Apr 12, 2019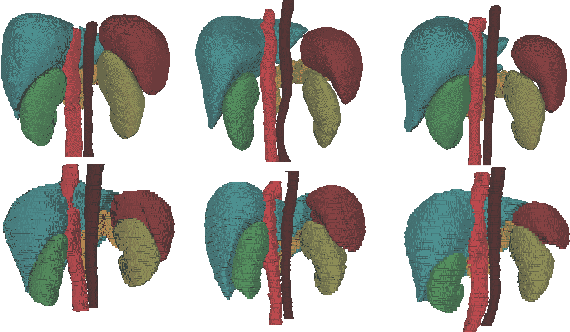
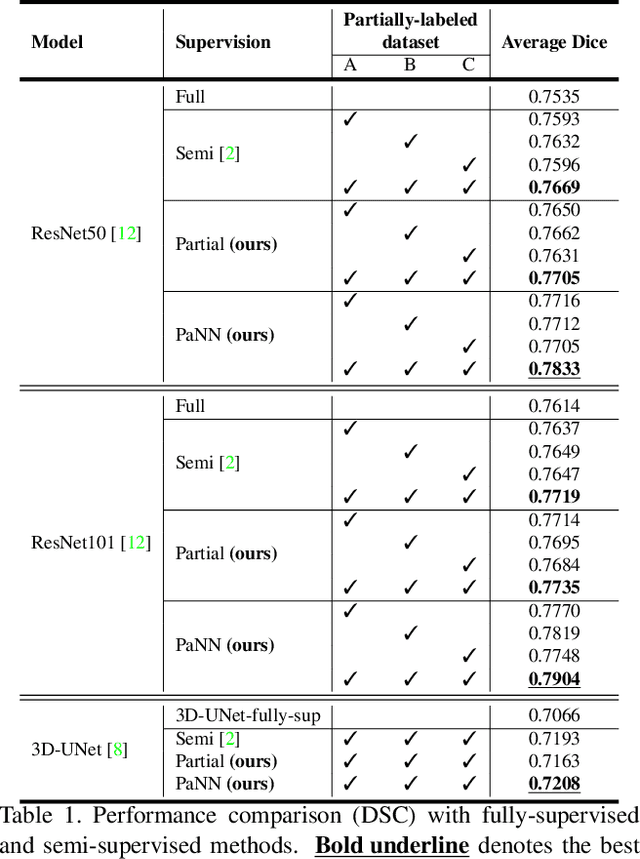
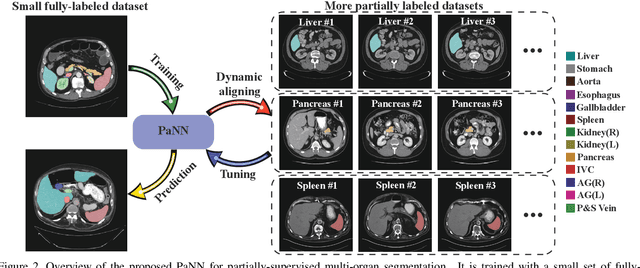
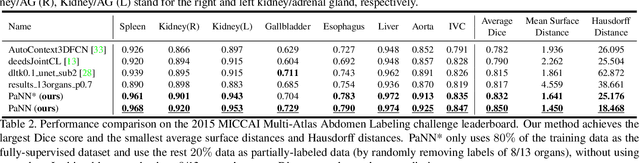
Abstract:Accurate multi-organ abdominal CT segmentation is essential to many clinical applications such as computer-aided intervention. As data annotation requires massive human labor from experienced radiologists, it is common that training data are partially labeled, e.g., pancreas datasets only have the pancreas labeled while leaving the rest marked as background. However, these background labels can be misleading in multi-organ segmentation since the "background" usually contains some other organs of interest. To address the background ambiguity in these partially-labeled datasets, we propose Prior-aware Neural Network (PaNN) via explicitly incorporating anatomical priors on abdominal organ sizes, guiding the training process with domain-specific knowledge. More specifically, PaNN assumes that the average organ size distributions in the abdomen should approximate their empirical distributions, a prior statistics obtained from the fully-labeled dataset. As our training objective is difficult to be directly optimized using stochastic gradient descent [20], we propose to reformulate it in a min-max form and optimize it via the stochastic primal-dual gradient algorithm. PaNN achieves state-of-the-art performance on the MICCAI2015 challenge "Multi-Atlas Labeling Beyond the Cranial Vault", a competition on organ segmentation in the abdomen. We report an average Dice score of 84.97%, surpassing the prior art by a large margin of 3.27%.
Abnormal Chest X-ray Identification With Generative Adversarial One-Class Classifier
Mar 05, 2019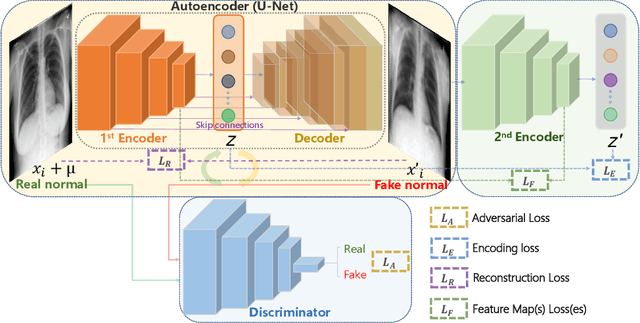
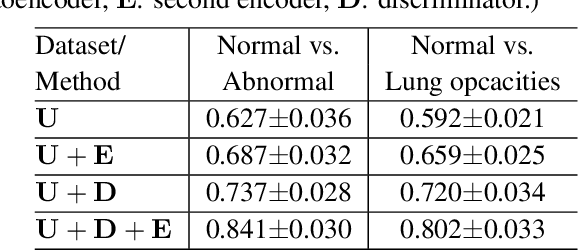
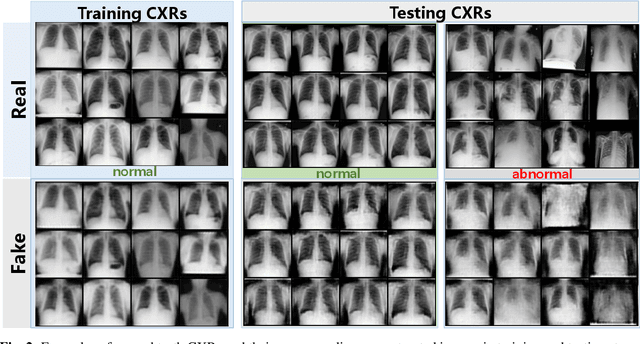
Abstract:Being one of the most common diagnostic imaging tests, chest radiography requires timely reporting of potential findings in the images. In this paper, we propose an end-to-end architecture for abnormal chest X-ray identification using generative adversarial one-class learning. Unlike previous approaches, our method takes only normal chest X-ray images as input. The architecture is composed of three deep neural networks, each of which learned by competing while collaborating among them to model the underlying content structure of the normal chest X-rays. Given a chest X-ray image in the testing phase, if it is normal, the learned architecture can well model and reconstruct the content; if it is abnormal, since the content is unseen in the training phase, the model would perform poorly in its reconstruction. It thus enables distinguishing abnormal chest X-rays from normal ones. Quantitative and qualitative experiments demonstrate the effectiveness and efficiency of our approach, where an AUC of 0.841 is achieved on the challenging NIH Chest X-ray dataset in a one-class learning setting, with the potential in reducing the workload for radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge