Kai Ding
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Beyond Conditional Computation: Retrieval-Augmented Genomic Foundation Models with Gengram
Jan 29, 2026Abstract:Current genomic foundation models (GFMs) rely on extensive neural computation to implicitly approximate conserved biological motifs from single-nucleotide inputs. We propose Gengram, a conditional memory module that introduces an explicit and highly efficient lookup primitive for multi-base motifs via a genomic-specific hashing scheme, establishing genomic "syntax". Integrated into the backbone of state-of-the-art GFMs, Gengram achieves substantial gains (up to 14%) across several functional genomics tasks. The module demonstrates robust architectural generalization, while further inspection of Gengram's latent space reveals the emergence of meaningful representations that align closely with fundamental biological knowledge. By establishing structured motif memory as a modeling primitive, Gengram simultaneously boosts empirical performance and mechanistic interpretability, providing a scalable and biology-aligned pathway for the next generation of GFMs. The code is available at https://github.com/zhejianglab/Genos, and the model checkpoint is available at https://huggingface.co/ZhejiangLab/Gengram.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Scale-Aware Curriculum Learning for Ddata-Efficient Lung Nodule Detection with YOLOv11
Oct 30, 2025Abstract:Lung nodule detection in chest CT is crucial for early lung cancer diagnosis, yet existing deep learning approaches face challenges when deployed in clinical settings with limited annotated data. While curriculum learning has shown promise in improving model training, traditional static curriculum strategies fail in data-scarce scenarios. We propose Scale Adaptive Curriculum Learning (SACL), a novel training strategy that dynamically adjusts curriculum design based on available data scale. SACL introduces three key mechanisms:(1) adaptive epoch scheduling, (2) hard sample injection, and (3) scale-aware optimization. We evaluate SACL on the LUNA25 dataset using YOLOv11 as the base detector. Experimental results demonstrate that while SACL achieves comparable performance to static curriculum learning on the full dataset in mAP50, it shows significant advantages under data-limited conditions with 4.6%, 3.5%, and 2.0% improvements over baseline at 10%, 20%, and 50% of training data respectively. By enabling robust training across varying data scales without architectural modifications, SACL provides a practical solution for healthcare institutions to develop effective lung nodule detection systems despite limited annotation resources.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
ProtoBERT-LoRA: Parameter-Efficient Prototypical Finetuning for Immunotherapy Study Identification
Mar 26, 2025Abstract:Identifying immune checkpoint inhibitor (ICI) studies in genomic repositories like Gene Expression Omnibus (GEO) is vital for cancer research yet remains challenging due to semantic ambiguity, extreme class imbalance, and limited labeled data in low-resource settings. We present ProtoBERT-LoRA, a hybrid framework that combines PubMedBERT with prototypical networks and Low-Rank Adaptation (LoRA) for efficient fine-tuning. The model enforces class-separable embeddings via episodic prototype training while preserving biomedical domain knowledge. Our dataset was divided as: Training (20 positive, 20 negative), Prototype Set (10 positive, 10 negative), Validation (20 positive, 200 negative), and Test (71 positive, 765 negative). Evaluated on test dataset, ProtoBERT-LoRA achieved F1-score of 0.624 (precision: 0.481, recall: 0.887), outperforming the rule-based system, machine learning baselines and finetuned PubMedBERT. Application to 44,287 unlabeled studies reduced manual review efforts by 82%. Ablation studies confirmed that combining prototypes with LoRA improved performance by 29% over stand-alone LoRA.
A Language Vision Model Approach for Automated Tumor Contouring in Radiation Oncology
Mar 19, 2025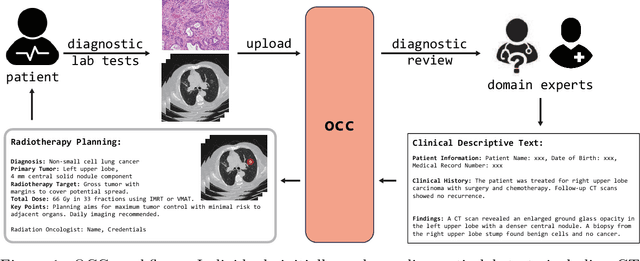
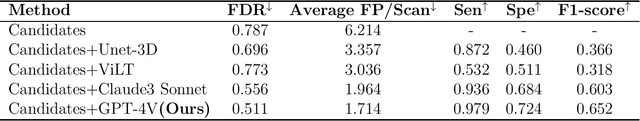
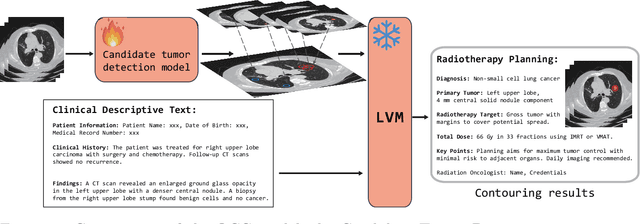
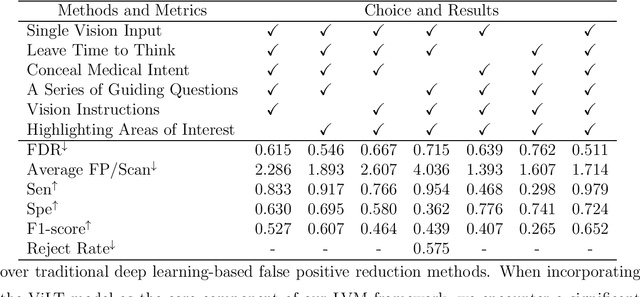
Abstract:Background: Lung cancer ranks as the leading cause of cancer-related mortality worldwide. The complexity of tumor delineation, crucial for radiation therapy, requires expertise often unavailable in resource-limited settings. Artificial Intelligence(AI), particularly with advancements in deep learning (DL) and natural language processing (NLP), offers potential solutions yet is challenged by high false positive rates. Purpose: The Oncology Contouring Copilot (OCC) system is developed to leverage oncologist expertise for precise tumor contouring using textual descriptions, aiming to increase the efficiency of oncological workflows by combining the strengths of AI with human oversight. Methods: Our OCC system initially identifies nodule candidates from CT scans. Employing Language Vision Models (LVMs) like GPT-4V, OCC then effectively reduces false positives with clinical descriptive texts, merging textual and visual data to automate tumor delineation, designed to elevate the quality of oncology care by incorporating knowledge from experienced domain experts. Results: Deployments of the OCC system resulted in a significant reduction in the false discovery rate by 35.0%, a 72.4% decrease in false positives per scan, and an F1-score of 0.652 across our dataset for unbiased evaluation. Conclusions: OCC represents a significant advance in oncology care, particularly through the use of the latest LVMs to improve contouring results by (1) streamlining oncology treatment workflows by optimizing tumor delineation, reducing manual processes; (2) offering a scalable and intuitive framework to reduce false positives in radiotherapy planning using LVMs; (3) introducing novel medical language vision prompt techniques to minimize LVMs hallucinations with ablation study, and (4) conducting a comparative analysis of LVMs, highlighting their potential in addressing medical language vision challenges.
Beyond Token Compression: A Training-Free Reduction Framework for Efficient Visual Processing in MLLMs
Jan 31, 2025



Abstract:Multimodal Large Language Models (MLLMs) are typically based on decoder-only or cross-attention architectures. While decoder-only MLLMs outperform their cross-attention counterparts, they require significantly higher computational resources due to extensive self-attention and FFN operations on visual tokens. This raises the question: can we eliminate these expensive operations while maintaining the performance? To this end, we present a novel analysis framework to investigate the necessity of these costly operations in decoder-only MLLMs. Our framework introduces two key innovations: (1) Hollow Attention, which limits visual token interactions to local attention while maintaining visual-text associations, and (2) Probe-Activated Dynamic FFN, which selectively activates FFN parameters for visual tokens. Both methods do not require fine-tuning, which significantly enhances analysis efficiency. To assess the impact of applying these reductions across different proportions of layers, we developed a greedy search method that significantly narrows the search space. Experiments on state-of-the-art MLLMs reveal that applying our reductions to approximately half of the layers not only maintains but sometimes improves model performance, indicating significant computational redundancy in current architectures. Additionally, our method is orthogonal to existing token compression techniques, allowing for further combination to achieve greater computational reduction. Our findings may provide valuable insights for the design of more efficient future MLLMs. Our code will be publicly available at https://github.com/L-Hugh/Beyond-Token-Compression.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025



Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Semantic Refocused Tuning for Open-Vocabulary Panoptic Segmentation
Sep 24, 2024



Abstract:Open-vocabulary panoptic segmentation is an emerging task aiming to accurately segment the image into semantically meaningful masks based on a set of texts. Despite existing efforts, it remains challenging to develop a high-performing method that generalizes effectively across new domains and requires minimal training resources. Our in-depth analysis of current methods reveals a crucial insight: mask classification is the main performance bottleneck for open-vocab. panoptic segmentation. Based on this, we propose Semantic Refocused Tuning (SMART), a novel framework that greatly enhances open-vocab. panoptic segmentation by improving mask classification through two key innovations. First, SMART adopts a multimodal Semantic-guided Mask Attention mechanism that injects task-awareness into the regional information extraction process. This enables the model to capture task-specific and contextually relevant information for more effective mask classification. Second, it incorporates Query Projection Tuning, which strategically fine-tunes the query projection layers within the Vision Language Model (VLM) used for mask classification. This adjustment allows the model to adapt the image focus of mask tokens to new distributions with minimal training resources, while preserving the VLM's pre-trained knowledge. Extensive ablation studies confirm the superiority of our approach. Notably, SMART sets new state-of-the-art results, demonstrating improvements of up to +1.3 PQ and +5.4 mIoU across representative benchmarks, while reducing training costs by nearly 10x compared to the previous best method. Our code and data will be released.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge