Xue Feng
University of California, Davis
Social World Model-Augmented Mechanism Design Policy Learning
Oct 22, 2025Abstract:Designing adaptive mechanisms to align individual and collective interests remains a central challenge in artificial social intelligence. Existing methods often struggle with modeling heterogeneous agents possessing persistent latent traits (e.g., skills, preferences) and dealing with complex multi-agent system dynamics. These challenges are compounded by the critical need for high sample efficiency due to costly real-world interactions. World Models, by learning to predict environmental dynamics, offer a promising pathway to enhance mechanism design in heterogeneous and complex systems. In this paper, we introduce a novel method named SWM-AP (Social World Model-Augmented Mechanism Design Policy Learning), which learns a social world model hierarchically modeling agents' behavior to enhance mechanism design. Specifically, the social world model infers agents' traits from their interaction trajectories and learns a trait-based model to predict agents' responses to the deployed mechanisms. The mechanism design policy collects extensive training trajectories by interacting with the social world model, while concurrently inferring agents' traits online during real-world interactions to further boost policy learning efficiency. Experiments in diverse settings (tax policy design, team coordination, and facility location) demonstrate that SWM-AP outperforms established model-based and model-free RL baselines in cumulative rewards and sample efficiency.
Cross-attention Secretly Performs Orthogonal Alignment in Recommendation Models
Oct 10, 2025Abstract:Cross-domain sequential recommendation (CDSR) aims to align heterogeneous user behavior sequences collected from different domains. While cross-attention is widely used to enhance alignment and improve recommendation performance, its underlying mechanism is not fully understood. Most researchers interpret cross-attention as residual alignment, where the output is generated by removing redundant and preserving non-redundant information from the query input by referencing another domain data which is input key and value. Beyond the prevailing view, we introduce Orthogonal Alignment, a phenomenon in which cross-attention discovers novel information that is not present in the query input, and further argue that those two contrasting alignment mechanisms can co-exist in recommendation models We find that when the query input and output of cross-attention are orthogonal, model performance improves over 300 experiments. Notably, Orthogonal Alignment emerges naturally, without any explicit orthogonality constraints. Our key insight is that Orthogonal Alignment emerges naturally because it improves scaling law. We show that baselines additionally incorporating cross-attention module outperform parameter-matched baselines, achieving a superior accuracy-per-model parameter. We hope these findings offer new directions for parameter-efficient scaling in multi-modal research.
Endoscopic Depth Estimation Based on Deep Learning: A Survey
Jul 28, 2025Abstract:Endoscopic depth estimation is a critical technology for improving the safety and precision of minimally invasive surgery. It has attracted considerable attention from researchers in medical imaging, computer vision, and robotics. Over the past decade, a large number of methods have been developed. Despite the existence of several related surveys, a comprehensive overview focusing on recent deep learning-based techniques is still limited. This paper endeavors to bridge this gap by systematically reviewing the state-of-the-art literature. Specifically, we provide a thorough survey of the field from three key perspectives: data, methods, and applications, covering a range of methods including both monocular and stereo approaches. We describe common performance evaluation metrics and summarize publicly available datasets. Furthermore, this review analyzes the specific challenges of endoscopic scenes and categorizes representative techniques based on their supervision strategies and network architectures. The application of endoscopic depth estimation in the important area of robot-assisted surgery is also reviewed. Finally, we outline potential directions for future research, such as domain adaptation, real-time implementation, and enhanced model generalization, thereby providing a valuable starting point for researchers to engage with and advance the field.
SIV-Bench: A Video Benchmark for Social Interaction Understanding and Reasoning
Jun 05, 2025Abstract:The rich and multifaceted nature of human social interaction, encompassing multimodal cues, unobservable relations and mental states, and dynamical behavior, presents a formidable challenge for artificial intelligence. To advance research in this area, we introduce SIV-Bench, a novel video benchmark for rigorously evaluating the capabilities of Multimodal Large Language Models (MLLMs) across Social Scene Understanding (SSU), Social State Reasoning (SSR), and Social Dynamics Prediction (SDP). SIV-Bench features 2,792 video clips and 8,792 meticulously generated question-answer pairs derived from a human-LLM collaborative pipeline. It is originally collected from TikTok and YouTube, covering a wide range of video genres, presentation styles, and linguistic and cultural backgrounds. It also includes a dedicated setup for analyzing the impact of different textual cues-original on-screen text, added dialogue, or no text. Our comprehensive experiments on leading MLLMs reveal that while models adeptly handle SSU, they significantly struggle with SSR and SDP, where Relation Inference (RI) is an acute bottleneck, as further examined in our analysis. Our study also confirms the critical role of transcribed dialogue in aiding comprehension of complex social interactions. By systematically identifying current MLLMs' strengths and limitations, SIV-Bench offers crucial insights to steer the development of more socially intelligent AI. The dataset and code are available at https://kfq20.github.io/sivbench/.
A Language Vision Model Approach for Automated Tumor Contouring in Radiation Oncology
Mar 19, 2025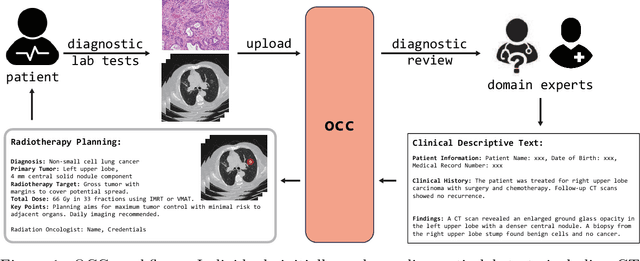
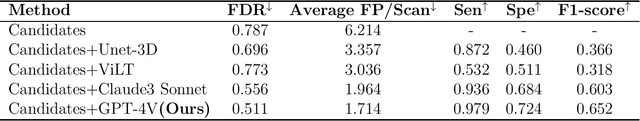
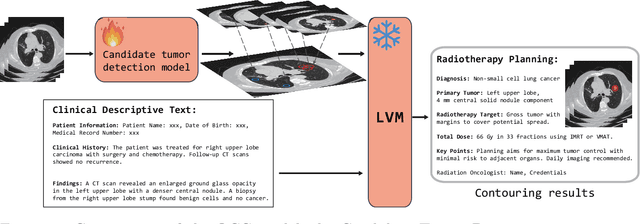
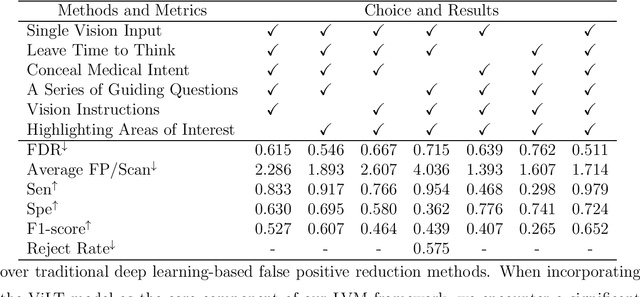
Abstract:Background: Lung cancer ranks as the leading cause of cancer-related mortality worldwide. The complexity of tumor delineation, crucial for radiation therapy, requires expertise often unavailable in resource-limited settings. Artificial Intelligence(AI), particularly with advancements in deep learning (DL) and natural language processing (NLP), offers potential solutions yet is challenged by high false positive rates. Purpose: The Oncology Contouring Copilot (OCC) system is developed to leverage oncologist expertise for precise tumor contouring using textual descriptions, aiming to increase the efficiency of oncological workflows by combining the strengths of AI with human oversight. Methods: Our OCC system initially identifies nodule candidates from CT scans. Employing Language Vision Models (LVMs) like GPT-4V, OCC then effectively reduces false positives with clinical descriptive texts, merging textual and visual data to automate tumor delineation, designed to elevate the quality of oncology care by incorporating knowledge from experienced domain experts. Results: Deployments of the OCC system resulted in a significant reduction in the false discovery rate by 35.0%, a 72.4% decrease in false positives per scan, and an F1-score of 0.652 across our dataset for unbiased evaluation. Conclusions: OCC represents a significant advance in oncology care, particularly through the use of the latest LVMs to improve contouring results by (1) streamlining oncology treatment workflows by optimizing tumor delineation, reducing manual processes; (2) offering a scalable and intuitive framework to reduce false positives in radiotherapy planning using LVMs; (3) introducing novel medical language vision prompt techniques to minimize LVMs hallucinations with ablation study, and (4) conducting a comparative analysis of LVMs, highlighting their potential in addressing medical language vision challenges.
FedOSAA: Improving Federated Learning with One-Step Anderson Acceleration
Mar 14, 2025



Abstract:Federated learning (FL) is a distributed machine learning approach that enables multiple local clients and a central server to collaboratively train a model while keeping the data on their own devices. First-order methods, particularly those incorporating variance reduction techniques, are the most widely used FL algorithms due to their simple implementation and stable performance. However, these methods tend to be slow and require a large number of communication rounds to reach the global minimizer. We propose FedOSAA, a novel approach that preserves the simplicity of first-order methods while achieving the rapid convergence typically associated with second-order methods. Our approach applies one Anderson acceleration (AA) step following classical local updates based on first-order methods with variance reduction, such as FedSVRG and SCAFFOLD, during local training. This AA step is able to leverage curvature information from the history points and gives a new update that approximates the Newton-GMRES direction, thereby significantly improving the convergence. We establish a local linear convergence rate to the global minimizer of FedOSAA for smooth and strongly convex loss functions. Numerical comparisons show that FedOSAA substantially improves the communication and computation efficiency of the original first-order methods, achieving performance comparable to second-order methods like GIANT.
ValuePilot: A Two-Phase Framework for Value-Driven Decision-Making
Mar 06, 2025



Abstract:Despite recent advances in artificial intelligence (AI), it poses challenges to ensure personalized decision-making in tasks that are not considered in training datasets. To address this issue, we propose ValuePilot, a two-phase value-driven decision-making framework comprising a dataset generation toolkit DGT and a decision-making module DMM trained on the generated data. DGT is capable of generating scenarios based on value dimensions and closely mirroring real-world tasks, with automated filtering techniques and human curation to ensure the validity of the dataset. In the generated dataset, DMM learns to recognize the inherent values of scenarios, computes action feasibility and navigates the trade-offs between multiple value dimensions to make personalized decisions. Extensive experiments demonstrate that, given human value preferences, our DMM most closely aligns with human decisions, outperforming Claude-3.5-Sonnet, Gemini-2-flash, Llama-3.1-405b and GPT-4o. This research is a preliminary exploration of value-driven decision-making. We hope it will stimulate interest in value-driven decision-making and personalized decision-making within the community.
Personalized Interpolation: An Efficient Method to Tame Flexible Optimization Window Estimation
Jan 23, 2025



Abstract:In the realm of online advertising, optimizing conversions is crucial for delivering relevant products to users and enhancing business outcomes. Predicting conversion events is challenging due to variable delays between user interactions, such as impressions or clicks, and the actual conversions. These delays differ significantly across various advertisers and products, necessitating distinct optimization time windows for targeted conversions. To address this, we introduce a novel approach named the \textit{Personalized Interpolation} method, which innovatively builds upon existing fixed conversion window models to estimate flexible conversion windows. This method allows for the accurate estimation of conversions across a variety of delay ranges, thus meeting the diverse needs of advertisers without increasing system complexity. To validate the efficacy of our proposed method, we conducted comprehensive experiments using ads conversion model. Our experiments demonstrate that this method not only achieves high prediction accuracy but also does so more efficiently than other existing solutions. This validation underscores the potential of our Personalized Interpolation method to significantly enhance conversion optimization in real-world online advertising systems, promising improved targeting and effectiveness in advertising strategies.
Causal Graph Guided Steering of LLM Values via Prompts and Sparse Autoencoders
Dec 31, 2024Abstract:As large language models (LLMs) become increasingly integrated into critical applications, aligning their behavior with human values presents significant challenges. Current methods, such as Reinforcement Learning from Human Feedback (RLHF), often focus on a limited set of values and can be resource-intensive. Furthermore, the correlation between values has been largely overlooked and remains underutilized. Our framework addresses this limitation by mining a causal graph that elucidates the implicit relationships among various values within the LLMs. Leveraging the causal graph, we implement two lightweight mechanisms for value steering: prompt template steering and Sparse Autoencoder feature steering, and analyze the effects of altering one value dimension on others. Extensive experiments conducted on Gemma-2B-IT and Llama3-8B-IT demonstrate the effectiveness and controllability of our steering methods.
Preference Discerning with LLM-Enhanced Generative Retrieval
Dec 11, 2024



Abstract:Sequential recommendation systems aim to provide personalized recommendations for users based on their interaction history. To achieve this, they often incorporate auxiliary information, such as textual descriptions of items and auxiliary tasks, like predicting user preferences and intent. Despite numerous efforts to enhance these models, they still suffer from limited personalization. To address this issue, we propose a new paradigm, which we term preference discerning. In preference dscerning, we explicitly condition a generative sequential recommendation system on user preferences within its context. To this end, we generate user preferences using Large Language Models (LLMs) based on user reviews and item-specific data. To evaluate preference discerning capabilities of sequential recommendation systems, we introduce a novel benchmark that provides a holistic evaluation across various scenarios, including preference steering and sentiment following. We assess current state-of-the-art methods using our benchmark and show that they struggle to accurately discern user preferences. Therefore, we propose a new method named Mender ($\textbf{M}$ultimodal Prefer$\textbf{en}$ce $\textbf{d}$iscern$\textbf{er}$), which improves upon existing methods and achieves state-of-the-art performance on our benchmark. Our results show that Mender can be effectively guided by human preferences even though they have not been observed during training, paving the way toward more personalized sequential recommendation systems. We will open-source the code and benchmarks upon publication.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge