Feng Shi
Mining fMRI Dynamics with Parcellation Prior for Brain Disease Diagnosis
May 04, 2023



Abstract:To characterize atypical brain dynamics under diseases, prevalent studies investigate functional magnetic resonance imaging (fMRI). However, most of the existing analyses compress rich spatial-temporal information as the brain functional networks (BFNs) and directly investigate the whole-brain network without neurological priors about functional subnetworks. We thus propose a novel graph learning framework to mine fMRI signals with topological priors from brain parcellation for disease diagnosis. Specifically, we 1) detect diagnosis-related temporal features using a "Transformer" for a higher-level BFN construction, and process it with a following graph convolutional network, and 2) apply an attention-based multiple instance learning strategy to emphasize the disease-affected subnetworks to further enhance the diagnosis performance and interpretability. Experiments demonstrate higher effectiveness of our method than compared methods in the diagnosis of early mild cognitive impairment. More importantly, our method is capable of localizing crucial brain subnetworks during the diagnosis, providing insights into the pathogenic source of mild cognitive impairment.
Self-supervised arbitrary scale super-resolution framework for anisotropic MRI
May 02, 2023Abstract:In this paper, we propose an efficient self-supervised arbitrary-scale super-resolution (SR) framework to reconstruct isotropic magnetic resonance (MR) images from anisotropic MRI inputs without involving external training data. The proposed framework builds a training dataset using in-the-wild anisotropic MR volumes with arbitrary image resolution. We then formulate the 3D volume SR task as a SR problem for 2D image slices. The anisotropic volume's high-resolution (HR) plane is used to build the HR-LR image pairs for model training. We further adapt the implicit neural representation (INR) network to implement the 2D arbitrary-scale image SR model. Finally, we leverage the well-trained proposed model to up-sample the 2D LR plane extracted from the anisotropic MR volumes to their HR views. The isotropic MR volumes thus can be reconstructed by stacking and averaging the generated HR slices. Our proposed framework has two major advantages: (1) It only involves the arbitrary-resolution anisotropic MR volumes, which greatly improves the model practicality in real MR imaging scenarios (e.g., clinical brain image acquisition); (2) The INR-based SR model enables arbitrary-scale image SR from the arbitrary-resolution input image, which significantly improves model training efficiency. We perform experiments on a simulated public adult brain dataset and a real collected 7T brain dataset. The results indicate that our current framework greatly outperforms two well-known self-supervised models for anisotropic MR image SR tasks.
Deep learning reveals the common spectrum underlying multiple brain disorders in youth and elders from brain functional networks
Feb 23, 2023



Abstract:Brain disorders in the early and late life of humans potentially share pathological alterations in brain functions. However, the key evidence from neuroimaging data for pathological commonness remains unrevealed. To explore this hypothesis, we build a deep learning model, using multi-site functional magnetic resonance imaging data (N=4,410, 6 sites), for classifying 5 different brain disorders from healthy controls, with a set of common features. Our model achieves 62.6(1.9)% overall classification accuracy on data from the 6 investigated sites and detects a set of commonly affected functional subnetworks at different spatial scales, including default mode, executive control, visual, and limbic networks. In the deep-layer feature representation for individual data, we observe young and aging patients with disorders are continuously distributed, which is in line with the clinical concept of the "spectrum of disorders". The revealed spectrum underlying early- and late-life brain disorders promotes the understanding of disorder comorbidities in the lifespan.
Single Cells Are Spatial Tokens: Transformers for Spatial Transcriptomic Data Imputation
Feb 06, 2023



Abstract:Spatially resolved transcriptomics brings exciting breakthroughs to single-cell analysis by providing physical locations along with gene expression. However, as a cost of the extremely high spatial resolution, the cellular level spatial transcriptomic data suffer significantly from missing values. While a standard solution is to perform imputation on the missing values, most existing methods either overlook spatial information or only incorporate localized spatial context without the ability to capture long-range spatial information. Using multi-head self-attention mechanisms and positional encoding, transformer models can readily grasp the relationship between tokens and encode location information. In this paper, by treating single cells as spatial tokens, we study how to leverage transformers to facilitate spatial tanscriptomics imputation. In particular, investigate the following two key questions: (1) $\textit{how to encode spatial information of cells in transformers}$, and (2) $\textit{ how to train a transformer for transcriptomic imputation}$. By answering these two questions, we present a transformer-based imputation framework, SpaFormer, for cellular-level spatial transcriptomic data. Extensive experiments demonstrate that SpaFormer outperforms existing state-of-the-art imputation algorithms on three large-scale datasets.
Runtime Performance of Evolutionary Algorithms for the Chance-constrained Makespan Scheduling Problem
Dec 22, 2022Abstract:The Makespan Scheduling problem is an extensively studied NP-hard problem, and its simplest version looks for an allocation approach for a set of jobs with deterministic processing times to two identical machines such that the makespan is minimized. However, in real life scenarios, the actual processing time of each job may be stochastic around the expected value with a variance, under the influence of external factors, and the actual processing times of these jobs may be correlated with covariances. Thus within this paper, we propose a chance-constrained version of the Makespan Scheduling problem and investigate the theoretical performance of the classical Randomized Local Search and (1+1) EA for it. More specifically, we first study two variants of the Chance-constrained Makespan Scheduling problem and their computational complexities, then separately analyze the expected runtime of the two algorithms to obtain an optimal solution or almost optimal solution to the instances of the two variants. In addition, we investigate the experimental performance of the two algorithms for the two variants.
Hierarchical Graph Convolutional Network Built by Multiscale Atlases for Brain Disorder Diagnosis Using Functional Connectivity
Sep 22, 2022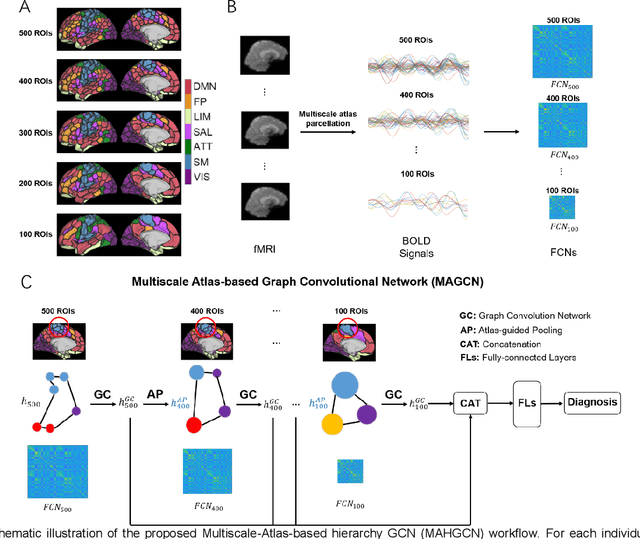
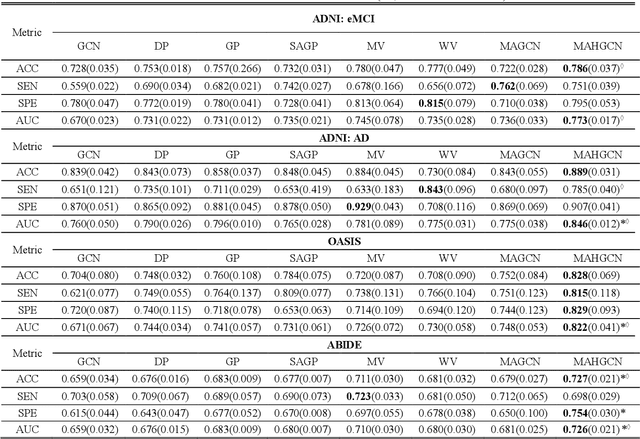
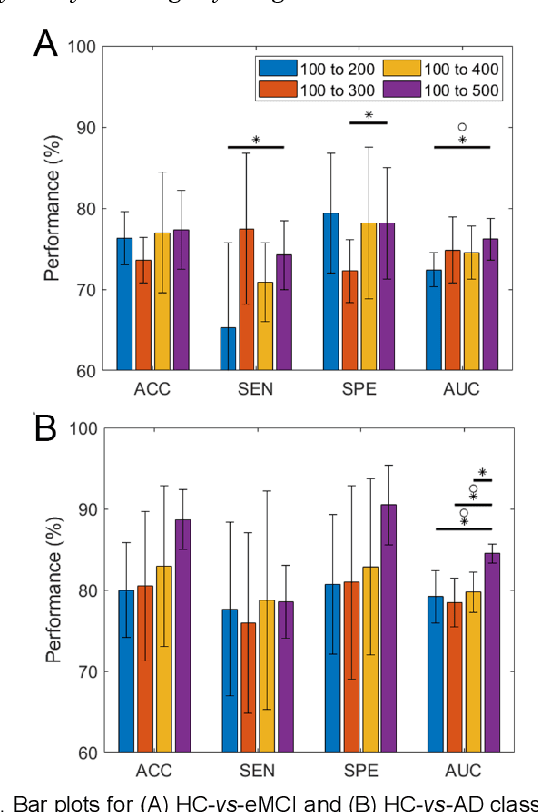
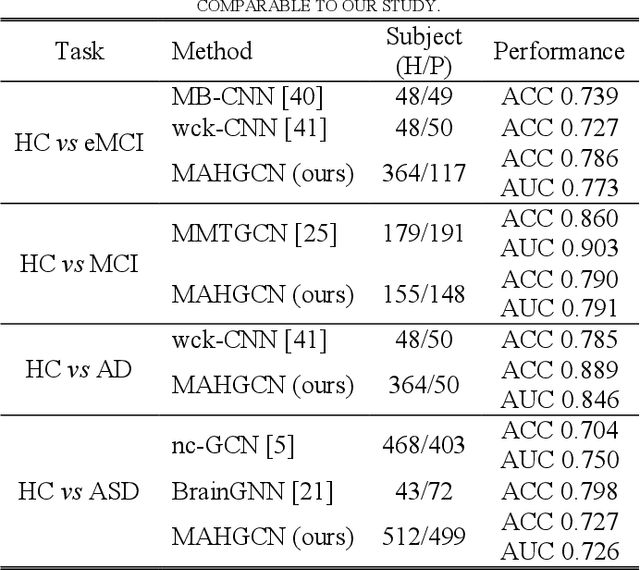
Abstract:Functional connectivity network (FCN) data from functional magnetic resonance imaging (fMRI) is increasingly used for the diagnoses of brain disorders. However, state-of-the-art studies used to build the FCN using a single brain parcellation atlas at a certain spatial scale, which largely neglected functional interactions across different spatial scales in hierarchical manners. In this study, we propose a novel framework to perform multiscale FCN analysis for brain disorder diagnosis. We first use a set of well-defined multiscale atlases to compute multiscale FCNs. Then, we utilize biologically meaningful brain hierarchical relationships among the regions in multiscale atlases to perform nodal pooling across multiple spatial scales, namely "Atlas-guided Pooling". Accordingly, we propose a Multiscale-Atlases-based Hierarchical Graph Convolutional Network (MAHGCN), built on the stacked layers of graph convolution and the atlas-guided pooling, for a comprehensive extraction of diagnostic information from multiscale FCNs. Experiments on neuroimaging data from 1792 subjects demonstrate the effectiveness of our proposed method in the diagnoses of Alzheimer's disease (AD), the prodromal stage of AD (i.e., mild cognitive impairment [MCI]), as well as autism spectrum disorder (ASD), with accuracy of 88.9%, 78.6%, and 72.7% respectively. All results show significant advantages of our proposed method over other competing methods. This study not only demonstrates the feasibility of brain disorder diagnosis using resting-state fMRI empowered by deep learning, but also highlights that the functional interactions in the multiscale brain hierarchy are worth being explored and integrated into deep learning network architectures for better understanding the neuropathology of brain disorders.
Alternately Optimized Graph Neural Networks
Jun 08, 2022
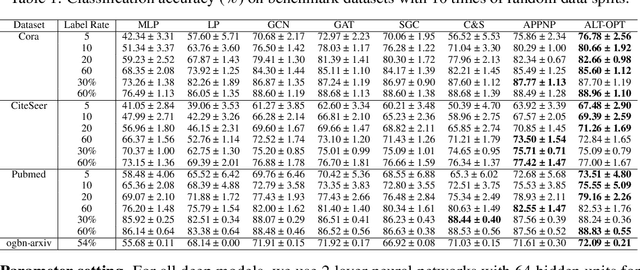

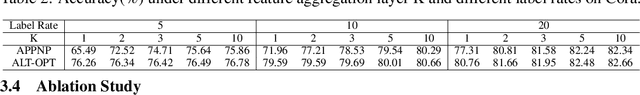
Abstract:Graph Neural Networks (GNNs) have demonstrated powerful representation capability in numerous graph-based tasks. Specifically, the decoupled structures of GNNs such as APPNP become popular due to their simplicity and performance advantages. However, the end-to-end training of these GNNs makes them inefficient in computation and memory consumption. In order to deal with these limitations, in this work, we propose an alternating optimization framework for graph neural networks that does not require end-to-end training. Extensive experiments under different settings demonstrate that the performance of the proposed algorithm is comparable to existing state-of-the-art algorithms but has significantly better computation and memory efficiency. Additionally, we show that our framework can be taken advantage to enhance existing decoupled GNNs.
Learning from the Tangram to Solve Mini Visual Tasks
Dec 12, 2021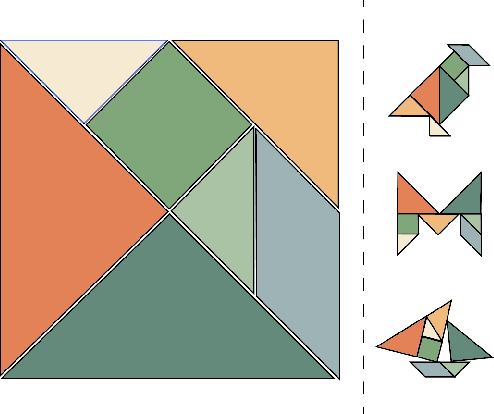
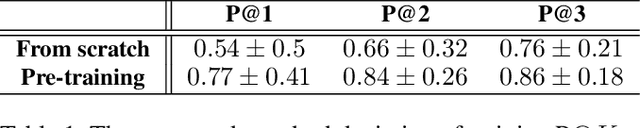
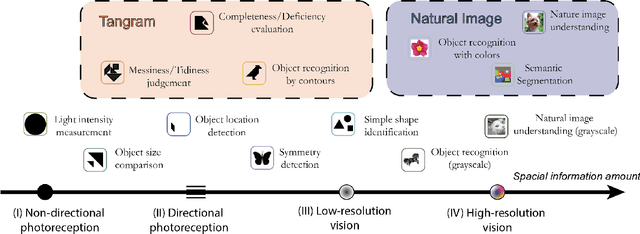
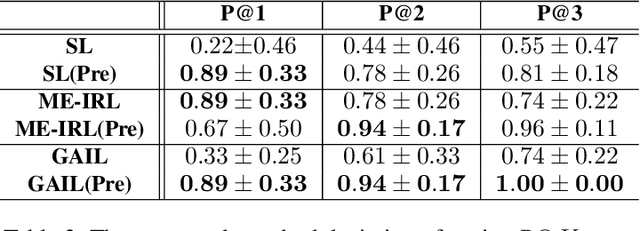
Abstract:Current pre-training methods in computer vision focus on natural images in the daily-life context. However, abstract diagrams such as icons and symbols are common and important in the real world. This work is inspired by Tangram, a game that requires replicating an abstract pattern from seven dissected shapes. By recording human experience in solving tangram puzzles, we present the Tangram dataset and show that a pre-trained neural model on the Tangram helps solve some mini visual tasks based on low-resolution vision. Extensive experiments demonstrate that our proposed method generates intelligent solutions for aesthetic tasks such as folding clothes and evaluating room layouts. The pre-trained feature extractor can facilitate the convergence of few-shot learning tasks on human handwriting and improve the accuracy in identifying icons by their contours. The Tangram dataset is available at https://github.com/yizhouzhao/Tangram.
Time Complexity Analysis of Evolutionary Algorithms for 2-Hop (1,2)-Minimum Spanning Tree Problem
Oct 10, 2021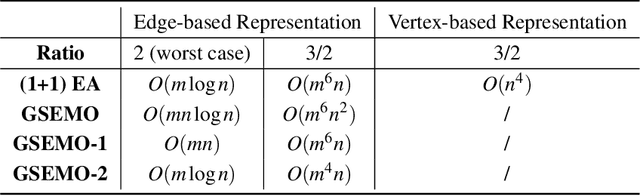
Abstract:The Minimum Spanning Tree problem (abbr. MSTP) is a well-known combinatorial optimization problem that has been extensively studied by the researchers in the field of evolutionary computing to theoretically analyze the optimization performance of evolutionary algorithms. Within the paper, we consider a constrained version of the problem named 2-Hop (1,2)-Minimum Spanning Tree problem (abbr. 2H-(1,2)-MSTP) in the context of evolutionary algorithms, which has been shown to be NP-hard. Following how evolutionary algorithms are applied to solve the MSTP, we first consider the evolutionary algorithms with search points in edge-based representation adapted to the 2H-(1,2)-MSTP (including the (1+1) EA, Global Simple Evolutionary Multi-Objective Optimizer and its two variants). More specifically, we separately investigate the upper bounds on their expected time (i.e., the expected number of fitness evaluations) to obtain a $\frac{3}{2}$-approximate solution with respect to different fitness functions. Inspired by the special structure of 2-hop spanning trees, we also consider the (1+1) EA with search points in vertex-based representation that seems not so natural for the problem and give an upper bound on its expected time to obtain a $\frac{3}{2}$-approximate solution, which is better than the above mentioned ones.
Cross-Site Severity Assessment of COVID-19 from CT Images via Domain Adaptation
Sep 08, 2021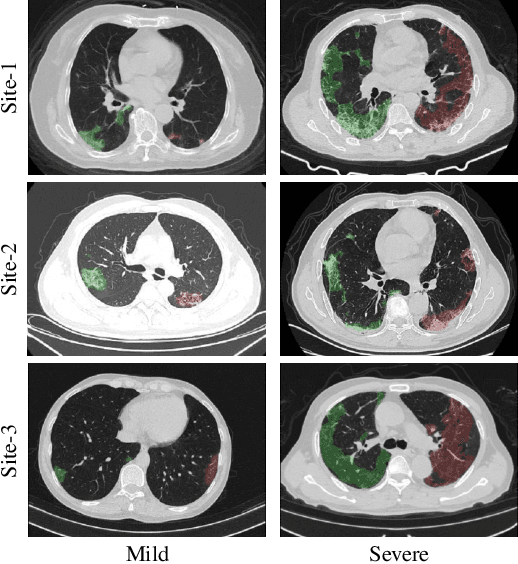
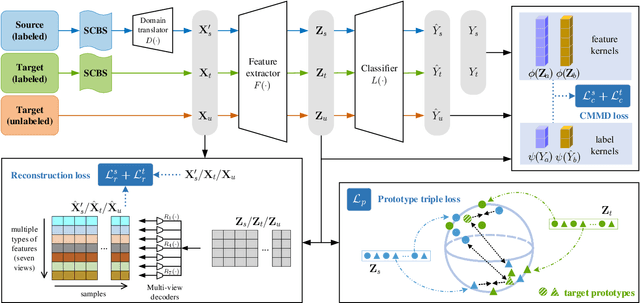
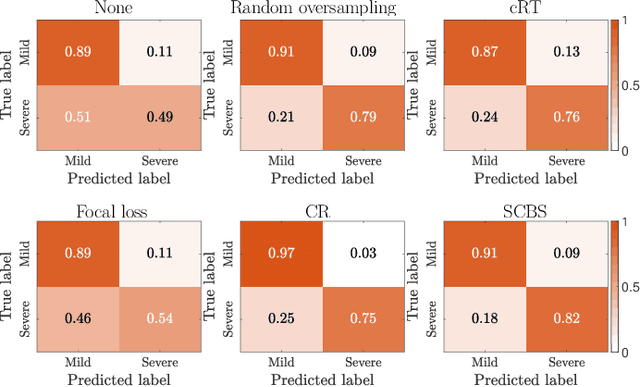
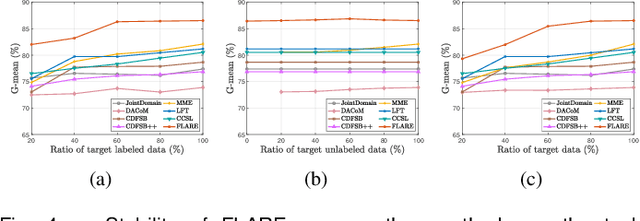
Abstract:Early and accurate severity assessment of Coronavirus disease 2019 (COVID-19) based on computed tomography (CT) images offers a great help to the estimation of intensive care unit event and the clinical decision of treatment planning. To augment the labeled data and improve the generalization ability of the classification model, it is necessary to aggregate data from multiple sites. This task faces several challenges including class imbalance between mild and severe infections, domain distribution discrepancy between sites, and presence of heterogeneous features. In this paper, we propose a novel domain adaptation (DA) method with two components to address these problems. The first component is a stochastic class-balanced boosting sampling strategy that overcomes the imbalanced learning problem and improves the classification performance on poorly-predicted classes. The second component is a representation learning that guarantees three properties: 1) domain-transferability by prototype triplet loss, 2) discriminant by conditional maximum mean discrepancy loss, and 3) completeness by multi-view reconstruction loss. Particularly, we propose a domain translator and align the heterogeneous data to the estimated class prototypes (i.e., class centers) in a hyper-sphere manifold. Experiments on cross-site severity assessment of COVID-19 from CT images show that the proposed method can effectively tackle the imbalanced learning problem and outperform recent DA approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge