Yuyao Zhang
Improving 2D Diffusion Models for 3D Medical Imaging with Inter-Slice Consistent Stochasticity
Feb 04, 2026Abstract:3D medical imaging is in high demand and essential for clinical diagnosis and scientific research. Currently, diffusion models (DMs) have become an effective tool for medical imaging reconstruction thanks to their ability to learn rich, high-quality data priors. However, learning the 3D data distribution with DMs in medical imaging is challenging, not only due to the difficulties in data collection but also because of the significant computational burden during model training. A common compromise is to train the DMs on 2D data priors and reconstruct stacked 2D slices to address 3D medical inverse problems. However, the intrinsic randomness of diffusion sampling causes severe inter-slice discontinuities of reconstructed 3D volumes. Existing methods often enforce continuity regularizations along the z-axis, which introduces sensitive hyper-parameters and may lead to over-smoothing results. In this work, we revisit the origin of stochasticity in diffusion sampling and introduce Inter-Slice Consistent Stochasticity (ISCS), a simple yet effective strategy that encourages interslice consistency during diffusion sampling. Our key idea is to control the consistency of stochastic noise components during diffusion sampling, thereby aligning their sampling trajectories without adding any new loss terms or optimization steps. Importantly, the proposed ISCS is plug-and-play and can be dropped into any 2D trained diffusion based 3D reconstruction pipeline without additional computational cost. Experiments on several medical imaging problems show that our method can effectively improve the performance of medical 3D imaging problems based on 2D diffusion models. Our findings suggest that controlling inter-slice stochasticity is a principled and practically attractive route toward high-fidelity 3D medical imaging with 2D diffusion priors. The code is available at: https://github.com/duchenhe/ISCS
Thinking-Based Non-Thinking: Solving the Reward Hacking Problem in Training Hybrid Reasoning Models via Reinforcement Learning
Jan 08, 2026Abstract:Large reasoning models (LRMs) have attracted much attention due to their exceptional performance. However, their performance mainly stems from thinking, a long Chain of Thought (CoT), which significantly increase computational overhead. To address this overthinking problem, existing work focuses on using reinforcement learning (RL) to train hybrid reasoning models that automatically decide whether to engage in thinking or not based on the complexity of the query. Unfortunately, using RL will suffer the the reward hacking problem, e.g., the model engages in thinking but is judged as not doing so, resulting in incorrect rewards. To mitigate this problem, existing works either employ supervised fine-tuning (SFT), which incurs high computational costs, or enforce uniform token limits on non-thinking responses, which yields limited mitigation of the problem. In this paper, we propose Thinking-Based Non-Thinking (TNT). It does not employ SFT, and sets different maximum token usage for responses not using thinking across various queries by leveraging information from the solution component of the responses using thinking. Experiments on five mathematical benchmarks demonstrate that TNT reduces token usage by around 50% compared to DeepSeek-R1-Distill-Qwen-1.5B/7B and DeepScaleR-1.5B, while significantly improving accuracy. In fact, TNT achieves the optimal trade-off between accuracy and efficiency among all tested methods. Additionally, the probability of reward hacking problem in TNT's responses, which are classified as not using thinking, remains below 10% across all tested datasets.
XDen-1K: A Density Field Dataset of Real-World Objects
Dec 11, 2025Abstract:A deep understanding of the physical world is a central goal for embodied AI and realistic simulation. While current models excel at capturing an object's surface geometry and appearance, they largely neglect its internal physical properties. This omission is critical, as properties like volumetric density are fundamental for predicting an object's center of mass, stability, and interaction dynamics in applications ranging from robotic manipulation to physical simulation. The primary bottleneck has been the absence of large-scale, real-world data. To bridge this gap, we introduce XDen-1K, the first large-scale, multi-modal dataset designed for real-world physical property estimation, with a particular focus on volumetric density. The core of this dataset consists of 1,000 real-world objects across 148 categories, for which we provide comprehensive multi-modal data, including a high-resolution 3D geometric model with part-level annotations and a corresponding set of real-world biplanar X-ray scans. Building upon this data, we introduce a novel optimization framework that recovers a high-fidelity volumetric density field of each object from its sparse X-ray views. To demonstrate its practical value, we add X-ray images as a conditioning signal to an existing segmentation network and perform volumetric segmentation. Furthermore, we conduct experiments on downstream robotics tasks. The results show that leveraging the dataset can effectively improve the accuracy of center-of-mass estimation and the success rate of robotic manipulation. We believe XDen-1K will serve as a foundational resource and a challenging new benchmark, catalyzing future research in physically grounded visual inference and embodied AI.
Diffusion Model Regularized Implicit Neural Representation for CT Metal Artifact Reduction
Dec 09, 2025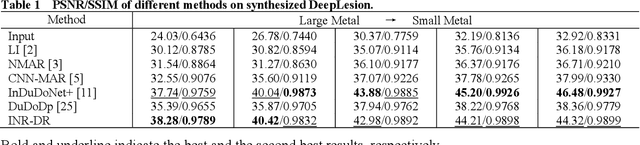
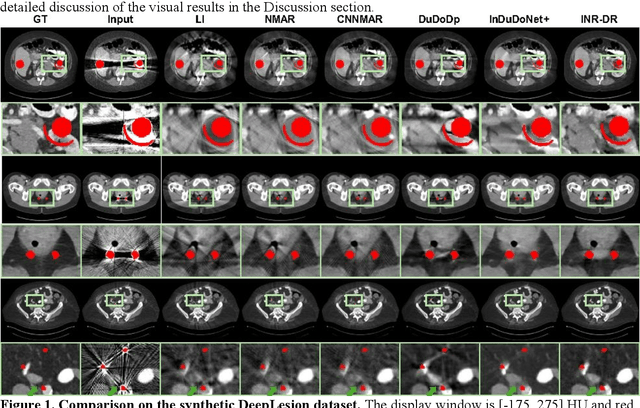
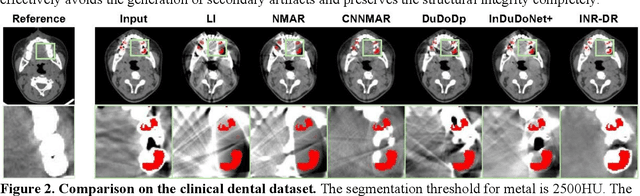
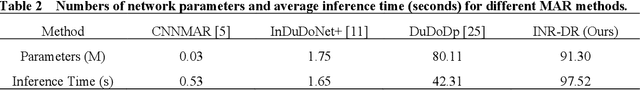
Abstract:Computed tomography (CT) images are often severely corrupted by artifacts in the presence of metals. Existing supervised metal artifact reduction (MAR) approaches suffer from performance instability on known data due to their reliance on limited paired metal-clean data, which limits their clinical applicability. Moreover, existing unsupervised methods face two main challenges: 1) the CT physical geometry is not effectively incorporated into the MAR process to ensure data fidelity; 2) traditional heuristics regularization terms cannot fully capture the abundant prior knowledge available. To overcome these shortcomings, we propose diffusion model regularized implicit neural representation framework for MAR. The implicit neural representation integrates physical constraints and imposes data fidelity, while the pre-trained diffusion model provides prior knowledge to regularize the solution. Experimental results on both simulated and clinical data demonstrate the effectiveness and generalization ability of our method, highlighting its potential to be applied to clinical settings.
Unsupervised Motion-Compensated Decomposition for Cardiac MRI Reconstruction via Neural Representation
Nov 17, 2025Abstract:Cardiac magnetic resonance (CMR) imaging is widely used to characterize cardiac morphology and function. To accelerate CMR imaging, various methods have been proposed to recover high-quality spatiotemporal CMR images from highly undersampled k-t space data. However, current CMR reconstruction techniques either fail to achieve satisfactory image quality or are restricted by the scarcity of ground truth data, leading to limited applicability in clinical scenarios. In this work, we proposed MoCo-INR, a new unsupervised method that integrates implicit neural representations (INR) with the conventional motion-compensated (MoCo) framework. Using explicit motion modeling and the continuous prior of INRs, MoCo-INR can produce accurate cardiac motion decomposition and high-quality CMR reconstruction. Furthermore, we introduce a new INR network architecture tailored to the CMR problem, which significantly stabilizes model optimization. Experiments on retrospective (simulated) datasets demonstrate the superiority of MoCo-INR over state-of-the-art methods, achieving fast convergence and fine-detailed reconstructions at ultra-high acceleration factors (e.g., 20x in VISTA sampling). Additionally, evaluations on prospective (real-acquired) free-breathing CMR scans highlight the clinical practicality of MoCo-INR for real-time imaging. Several ablation studies further confirm the effectiveness of the critical components of MoCo-INR.
Decoupled Planning and Execution: A Hierarchical Reasoning Framework for Deep Search
Jul 03, 2025Abstract:Complex information needs in real-world search scenarios demand deep reasoning and knowledge synthesis across diverse sources, which traditional retrieval-augmented generation (RAG) pipelines struggle to address effectively. Current reasoning-based approaches suffer from a fundamental limitation: they use a single model to handle both high-level planning and detailed execution, leading to inefficient reasoning and limited scalability. In this paper, we introduce HiRA, a hierarchical framework that separates strategic planning from specialized execution. Our approach decomposes complex search tasks into focused subtasks, assigns each subtask to domain-specific agents equipped with external tools and reasoning capabilities, and coordinates the results through a structured integration mechanism. This separation prevents execution details from disrupting high-level reasoning while enabling the system to leverage specialized expertise for different types of information processing. Experiments on four complex, cross-modal deep search benchmarks demonstrate that HiRA significantly outperforms state-of-the-art RAG and agent-based systems. Our results show improvements in both answer quality and system efficiency, highlighting the effectiveness of decoupled planning and execution for multi-step information seeking tasks. Our code is available at https://github.com/ignorejjj/HiRA.
Leveraging LLM-Assisted Query Understanding for Live Retrieval-Augmented Generation
Jun 26, 2025


Abstract:Real-world live retrieval-augmented generation (RAG) systems face significant challenges when processing user queries that are often noisy, ambiguous, and contain multiple intents. While RAG enhances large language models (LLMs) with external knowledge, current systems typically struggle with such complex inputs, as they are often trained or evaluated on cleaner data. This paper introduces Omni-RAG, a novel framework designed to improve the robustness and effectiveness of RAG systems in live, open-domain settings. Omni-RAG employs LLM-assisted query understanding to preprocess user inputs through three key modules: (1) Deep Query Understanding and Decomposition, which utilizes LLMs with tailored prompts to denoise queries (e.g., correcting spelling errors) and decompose multi-intent queries into structured sub-queries; (2) Intent-Aware Knowledge Retrieval, which performs retrieval for each sub-query from a corpus (i.e., FineWeb using OpenSearch) and aggregates the results; and (3) Reranking and Generation, where a reranker (i.e., BGE) refines document selection before a final response is generated by an LLM (i.e., Falcon-10B) using a chain-of-thought prompt. Omni-RAG aims to bridge the gap between current RAG capabilities and the demands of real-world applications, such as those highlighted by the SIGIR 2025 LiveRAG Challenge, by robustly handling complex and noisy queries.
Low-Rank Augmented Implicit Neural Representation for Unsupervised High-Dimensional Quantitative MRI Reconstruction
Jun 10, 2025Abstract:Quantitative magnetic resonance imaging (qMRI) provides tissue-specific parameters vital for clinical diagnosis. Although simultaneous multi-parametric qMRI (MP-qMRI) technologies enhance imaging efficiency, robustly reconstructing qMRI from highly undersampled, high-dimensional measurements remains a significant challenge. This difficulty arises primarily because current reconstruction methods that rely solely on a single prior or physics-informed model to solve the highly ill-posed inverse problem, which often leads to suboptimal results. To overcome this limitation, we propose LoREIN, a novel unsupervised and dual-prior-integrated framework for accelerated 3D MP-qMRI reconstruction. Technically, LoREIN incorporates both low-rank prior and continuity prior via low-rank representation (LRR) and implicit neural representation (INR), respectively, to enhance reconstruction fidelity. The powerful continuous representation of INR enables the estimation of optimal spatial bases within the low-rank subspace, facilitating high-fidelity reconstruction of weighted images. Simultaneously, the predicted multi-contrast weighted images provide essential structural and quantitative guidance, further enhancing the reconstruction accuracy of quantitative parameter maps. Furthermore, our work introduces a zero-shot learning paradigm with broad potential in complex spatiotemporal and high-dimensional image reconstruction tasks, further advancing the field of medical imaging.
SmartAvatar: Text- and Image-Guided Human Avatar Generation with VLM AI Agents
Jun 05, 2025



Abstract:SmartAvatar is a vision-language-agent-driven framework for generating fully rigged, animation-ready 3D human avatars from a single photo or textual prompt. While diffusion-based methods have made progress in general 3D object generation, they continue to struggle with precise control over human identity, body shape, and animation readiness. In contrast, SmartAvatar leverages the commonsense reasoning capabilities of large vision-language models (VLMs) in combination with off-the-shelf parametric human generators to deliver high-quality, customizable avatars. A key innovation is an autonomous verification loop, where the agent renders draft avatars, evaluates facial similarity, anatomical plausibility, and prompt alignment, and iteratively adjusts generation parameters for convergence. This interactive, AI-guided refinement process promotes fine-grained control over both facial and body features, enabling users to iteratively refine their avatars via natural-language conversations. Unlike diffusion models that rely on static pre-trained datasets and offer limited flexibility, SmartAvatar brings users into the modeling loop and ensures continuous improvement through an LLM-driven procedural generation and verification system. The generated avatars are fully rigged and support pose manipulation with consistent identity and appearance, making them suitable for downstream animation and interactive applications. Quantitative benchmarks and user studies demonstrate that SmartAvatar outperforms recent text- and image-driven avatar generation systems in terms of reconstructed mesh quality, identity fidelity, attribute accuracy, and animation readiness, making it a versatile tool for realistic, customizable avatar creation on consumer-grade hardware.
SUFFICIENT: A scan-specific unsupervised deep learning framework for high-resolution 3D isotropic fetal brain MRI reconstruction
May 26, 2025Abstract:High-quality 3D fetal brain MRI reconstruction from motion-corrupted 2D slices is crucial for clinical diagnosis. Reliable slice-to-volume registration (SVR)-based motion correction and super-resolution reconstruction (SRR) methods are essential. Deep learning (DL) has demonstrated potential in enhancing SVR and SRR when compared to conventional methods. However, it requires large-scale external training datasets, which are difficult to obtain for clinical fetal MRI. To address this issue, we propose an unsupervised iterative SVR-SRR framework for isotropic HR volume reconstruction. Specifically, SVR is formulated as a function mapping a 2D slice and a 3D target volume to a rigid transformation matrix, which aligns the slice to the underlying location in the target volume. The function is parameterized by a convolutional neural network, which is trained by minimizing the difference between the volume slicing at the predicted position and the input slice. In SRR, a decoding network embedded within a deep image prior framework is incorporated with a comprehensive image degradation model to produce the high-resolution (HR) volume. The deep image prior framework offers a local consistency prior to guide the reconstruction of HR volumes. By performing a forward degradation model, the HR volume is optimized by minimizing loss between predicted slices and the observed slices. Comprehensive experiments conducted on large-magnitude motion-corrupted simulation data and clinical data demonstrate the superior performance of the proposed framework over state-of-the-art fetal brain reconstruction frameworks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge