Shilei Cao
Unveiling Modality Bias: Automated Sample-Specific Analysis for Multimodal Misinformation Benchmarks
Nov 08, 2025Abstract:Numerous multimodal misinformation benchmarks exhibit bias toward specific modalities, allowing detectors to make predictions based solely on one modality. While previous research has quantified bias at the dataset level or manually identified spurious correlations between modalities and labels, these approaches lack meaningful insights at the sample level and struggle to scale to the vast amount of online information. In this paper, we investigate the design for automated recognition of modality bias at the sample level. Specifically, we propose three bias quantification methods based on theories/views of different levels of granularity: 1) a coarse-grained evaluation of modality benefit; 2) a medium-grained quantification of information flow; and 3) a fine-grained causality analysis. To verify the effectiveness, we conduct a human evaluation on two popular benchmarks. Experimental results reveal three interesting findings that provide potential direction toward future research: 1)~Ensembling multiple views is crucial for reliable automated analysis; 2)~Automated analysis is prone to detector-induced fluctuations; and 3)~Different views produce a higher agreement on modality-balanced samples but diverge on biased ones.
Task-Adaptive Parameter-Efficient Fine-Tuning for Weather Foundation Models
Sep 26, 2025Abstract:While recent advances in machine learning have equipped Weather Foundation Models (WFMs) with substantial generalization capabilities across diverse downstream tasks, the escalating computational requirements associated with their expanding scale increasingly hinder practical deployment. Current Parameter-Efficient Fine-Tuning (PEFT) methods, designed for vision or language tasks, fail to address the unique challenges of weather downstream tasks, such as variable heterogeneity, resolution diversity, and spatiotemporal coverage variations, leading to suboptimal performance when applied to WFMs. To bridge this gap, we introduce WeatherPEFT, a novel PEFT framework for WFMs incorporating two synergistic innovations. First, during the forward pass, Task-Adaptive Dynamic Prompting (TADP) dynamically injects the embedding weights within the encoder to the input tokens of the pre-trained backbone via internal and external pattern extraction, enabling context-aware feature recalibration for specific downstream tasks. Furthermore, during backpropagation, Stochastic Fisher-Guided Adaptive Selection (SFAS) not only leverages Fisher information to identify and update the most task-critical parameters, thereby preserving invariant pre-trained knowledge, but also introduces randomness to stabilize the selection. We demonstrate the effectiveness and efficiency of WeatherPEFT on three downstream tasks, where existing PEFT methods show significant gaps versus Full-Tuning, and WeatherPEFT achieves performance parity with Full-Tuning using fewer trainable parameters. The code of this work will be released.
TianQuan-Climate: A Subseasonal-to-Seasonal Global Weather Model via Incorporate Climatology State
Apr 14, 2025Abstract:Subseasonal forecasting serves as an important support for Sustainable Development Goals (SDGs), such as climate challenges, agricultural yield and sustainable energy production. However, subseasonal forecasting is a complex task in meteorology due to dissipating initial conditions and delayed external forces. Although AI models are increasingly pushing the boundaries of this forecasting limit, they face two major challenges: error accumulation and Smoothness. To address these two challenges, we propose Climate Furnace Subseasonal-to-Seasonal (TianQuan-Climate), a novel machine learning model designed to provide global daily mean forecasts up to 45 days, covering five upper-air atmospheric variables at 13 pressure levels and two surface variables. Our proposed TianQuan-Climate has two advantages: 1) it utilizes a multi-model prediction strategy to reduce system error impacts in long-term subseasonal forecasts; 2) it incorporates a Content Fusion Module for climatological integration and extends ViT with uncertainty blocks (UD-ViT) to improve generalization by learning from uncertainty. We demonstrate the effectiveness of TianQuan-Climate on benchmarks for weather forecasting and climate projections within the 15 to 45-day range, where TianQuan-Climate outperforms existing numerical and AI methods.
Exploring Test-Time Adaptation for Object Detection in Continually Changing Environments
Jun 25, 2024Abstract:For real-world applications, neural network models are commonly deployed in dynamic environments, where the distribution of the target domain undergoes temporal changes. Continual Test-Time Adaptation (CTTA) has recently emerged as a promising technique to gradually adapt a source-trained model to test data drawn from a continually changing target domain. Despite recent advancements in addressing CTTA, two critical issues remain: 1) The use of a fixed threshold for pseudo-labeling in existing methodologies leads to the generation of low-quality pseudo-labels, as model confidence varies across categories and domains; 2) While current solutions utilize stochastic parameter restoration to mitigate catastrophic forgetting, their capacity to preserve critical information is undermined by its intrinsic randomness. To tackle these challenges, we present CTAOD, aiming to enhance the performance of detection models in CTTA scenarios. Inspired by prior CTTA works for effective adaptation, CTAOD is founded on the mean-teacher framework, characterized by three core components. Firstly, the object-level contrastive learning module tailored for object detection extracts object-level features using the teacher's region of interest features and optimizes them through contrastive learning. Secondly, the dynamic threshold strategy updates the category-specific threshold based on predicted confidence scores to improve the quality of pseudo-labels. Lastly, we design a data-driven stochastic restoration mechanism to selectively reset inactive parameters using the gradients as weights for a random mask matrix, thereby ensuring the retention of essential knowledge. We demonstrate the effectiveness of our approach on four CTTA tasks for object detection, where CTAOD outperforms existing methods, especially achieving a 3.0 mAP improvement on the Cityscapes-to-Cityscapes-C CTTA task.
A$^{2}$-MAE: A spatial-temporal-spectral unified remote sensing pre-training method based on anchor-aware masked autoencoder
Jun 12, 2024



Abstract:Vast amounts of remote sensing (RS) data provide Earth observations across multiple dimensions, encompassing critical spatial, temporal, and spectral information which is essential for addressing global-scale challenges such as land use monitoring, disaster prevention, and environmental change mitigation. Despite various pre-training methods tailored to the characteristics of RS data, a key limitation persists: the inability to effectively integrate spatial, temporal, and spectral information within a single unified model. To unlock the potential of RS data, we construct a Spatial-Temporal-Spectral Structured Dataset (STSSD) characterized by the incorporation of multiple RS sources, diverse coverage, unified locations within image sets, and heterogeneity within images. Building upon this structured dataset, we propose an Anchor-Aware Masked AutoEncoder method (A$^{2}$-MAE), leveraging intrinsic complementary information from the different kinds of images and geo-information to reconstruct the masked patches during the pre-training phase. A$^{2}$-MAE integrates an anchor-aware masking strategy and a geographic encoding module to comprehensively exploit the properties of RS images. Specifically, the proposed anchor-aware masking strategy dynamically adapts the masking process based on the meta-information of a pre-selected anchor image, thereby facilitating the training on images captured by diverse types of RS sources within one model. Furthermore, we propose a geographic encoding method to leverage accurate spatial patterns, enhancing the model generalization capabilities for downstream applications that are generally location-related. Extensive experiments demonstrate our method achieves comprehensive improvements across various downstream tasks compared with existing RS pre-training methods, including image classification, semantic segmentation, and change detection tasks.
Learning Shape Priors by Pairwise Comparison for Robust Semantic Segmentation
Apr 23, 2022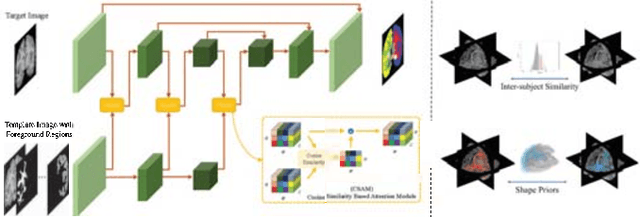
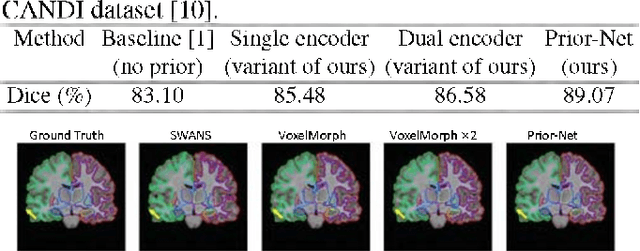
Abstract:Semantic segmentation is important in medical image analysis. Inspired by the strong ability of traditional image analysis techniques in capturing shape priors and inter-subject similarity, many deep learning (DL) models have been recently proposed to exploit such prior information and achieved robust performance. However, these two types of important prior information are usually studied separately in existing models. In this paper, we propose a novel DL model to model both type of priors within a single framework. Specifically, we introduce an extra encoder into the classic encoder-decoder structure to form a Siamese structure for the encoders, where one of them takes a target image as input (the image-encoder), and the other concatenates a template image and its foreground regions as input (the template-encoder). The template-encoder encodes the shape priors and appearance characteristics of each foreground class in the template image. A cosine similarity based attention module is proposed to fuse the information from both encoders, to utilize both types of prior information encoded by the template-encoder and model the inter-subject similarity for each foreground class. Extensive experiments on two public datasets demonstrate that our proposed method can produce superior performance to competing methods.
Conquering Data Variations in Resolution: A Slice-Aware Multi-Branch Decoder Network
Mar 07, 2022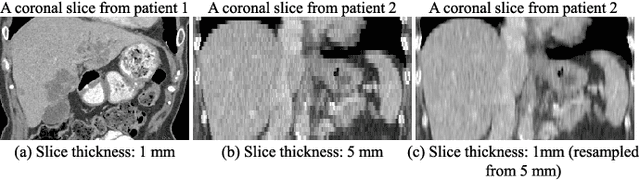
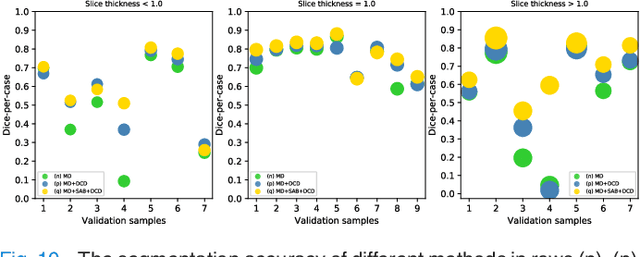
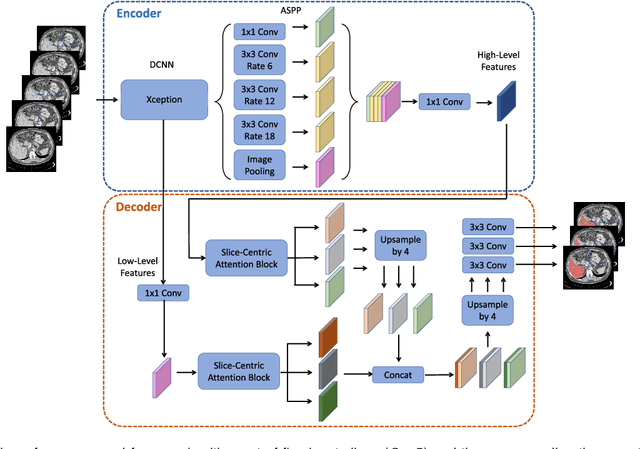
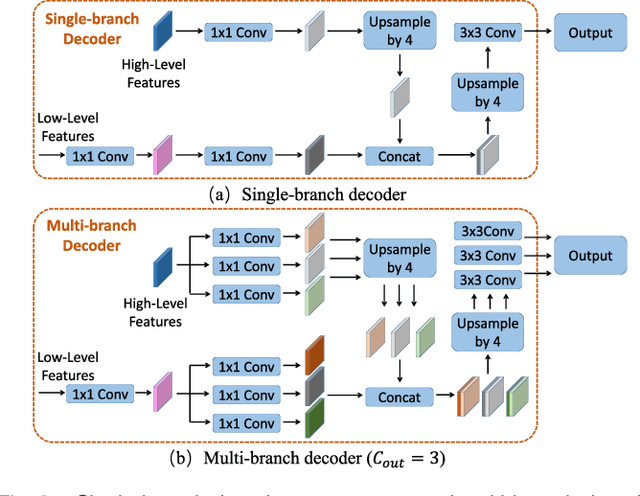
Abstract:Fully convolutional neural networks have made promising progress in joint liver and liver tumor segmentation. Instead of following the debates over 2D versus 3D networks (for example, pursuing the balance between large-scale 2D pretraining and 3D context), in this paper, we novelly identify the wide variation in the ratio between intra- and inter-slice resolutions as a crucial obstacle to the performance. To tackle the mismatch between the intra- and inter-slice information, we propose a slice-aware 2.5D network that emphasizes extracting discriminative features utilizing not only in-plane semantics but also out-of-plane coherence for each separate slice. Specifically, we present a slice-wise multi-input multi-output architecture to instantiate such a design paradigm, which contains a Multi-Branch Decoder (MD) with a Slice-centric Attention Block (SAB) for learning slice-specific features and a Densely Connected Dice (DCD) loss to regularize the inter-slice predictions to be coherent and continuous. Based on the aforementioned innovations, we achieve state-of-the-art results on the MICCAI 2017 Liver Tumor Segmentation (LiTS) dataset. Besides, we also test our model on the ISBI 2019 Segmentation of THoracic Organs at Risk (SegTHOR) dataset, and the result proves the robustness and generalizability of the proposed method in other segmentation tasks.
RECIST-Net: Lesion detection via grouping keypoints on RECIST-based annotation
Jul 19, 2021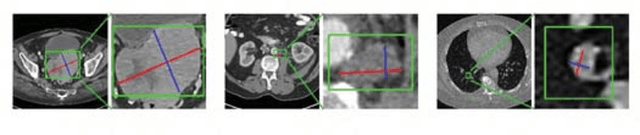
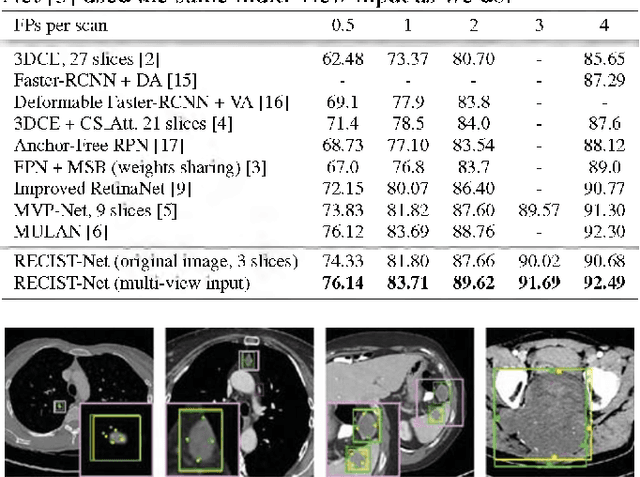
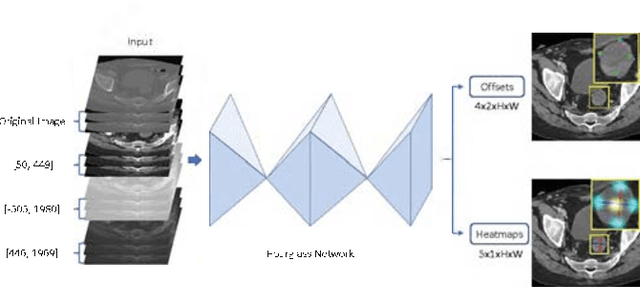
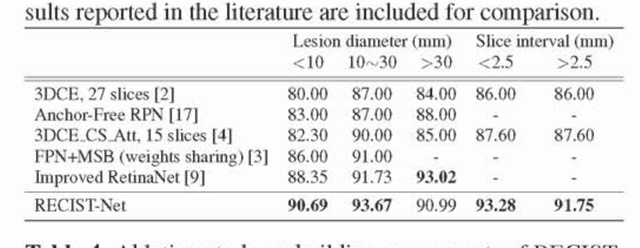
Abstract:Universal lesion detection in computed tomography (CT) images is an important yet challenging task due to the large variations in lesion type, size, shape, and appearance. Considering that data in clinical routine (such as the DeepLesion dataset) are usually annotated with a long and a short diameter according to the standard of Response Evaluation Criteria in Solid Tumors (RECIST) diameters, we propose RECIST-Net, a new approach to lesion detection in which the four extreme points and center point of the RECIST diameters are detected. By detecting a lesion as keypoints, we provide a more conceptually straightforward formulation for detection, and overcome several drawbacks (e.g., requiring extensive effort in designing data-appropriate anchors and losing shape information) of existing bounding-box-based methods while exploring a single-task, one-stage approach compared to other RECIST-based approaches. Experiments show that RECIST-Net achieves a sensitivity of 92.49% at four false positives per image, outperforming other recent methods including those using multi-task learning.
Generalized Organ Segmentation by Imitating One-shot Reasoning using Anatomical Correlation
Mar 30, 2021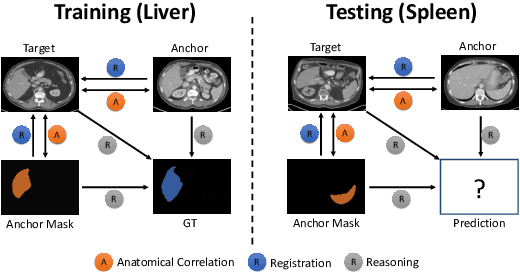
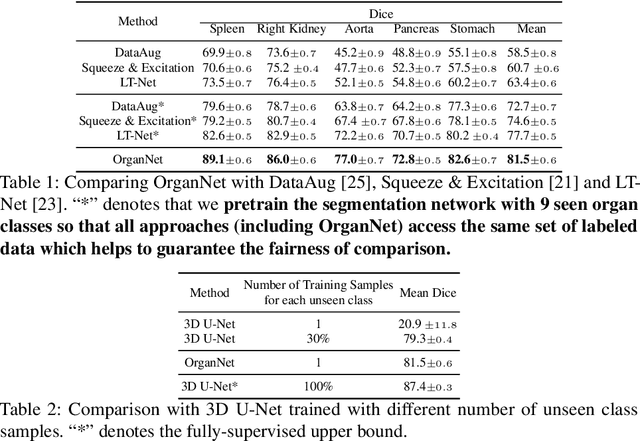
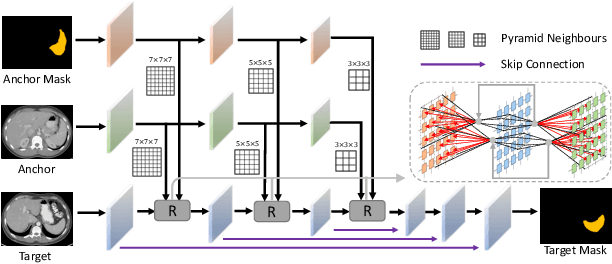
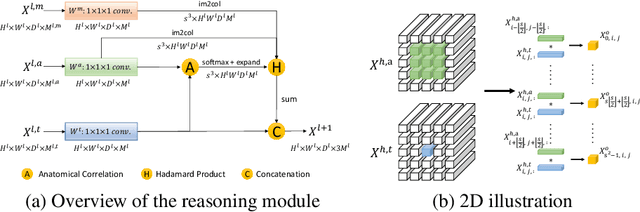
Abstract:Learning by imitation is one of the most significant abilities of human beings and plays a vital role in human's computational neural system. In medical image analysis, given several exemplars (anchors), experienced radiologist has the ability to delineate unfamiliar organs by imitating the reasoning process learned from existing types of organs. Inspired by this observation, we propose OrganNet which learns a generalized organ concept from a set of annotated organ classes and then transfer this concept to unseen classes. In this paper, we show that such process can be integrated into the one-shot segmentation task which is a very challenging but meaningful topic. We propose pyramid reasoning modules (PRMs) to model the anatomical correlation between anchor and target volumes. In practice, the proposed module first computes a correlation matrix between target and anchor computerized tomography (CT) volumes. Then, this matrix is used to transform the feature representations of both anchor volume and its segmentation mask. Finally, OrganNet learns to fuse the representations from various inputs and predicts segmentation results for target volume. Extensive experiments show that OrganNet can effectively resist the wide variations in organ morphology and produce state-of-the-art results in one-shot segmentation task. Moreover, even when compared with fully-supervised segmentation models, OrganNet is still able to produce satisfying segmentation results.
Brain Atlas Guided Attention U-Net for White Matter Hyperintensity Segmentation
Oct 19, 2020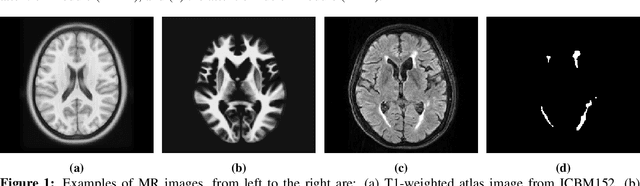

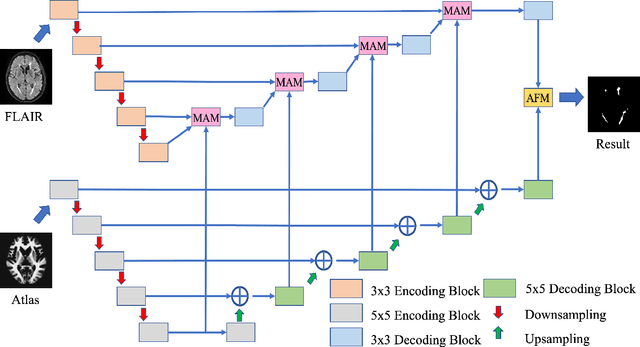
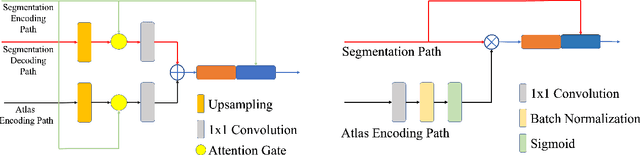
Abstract:White Matter Hyperintensities (WMH) are the most common manifestation of cerebral small vessel disease (cSVD) on the brain MRI. Accurate WMH segmentation algorithms are important to determine cSVD burden and its clinical consequences. Most of existing WMH segmentation algorithms require both fluid attenuated inversion recovery (FLAIR) images and T1-weighted images as inputs. However, T1-weighted images are typically not part of standard clinicalscans which are acquired for patients with acute stroke. In this paper, we propose a novel brain atlas guided attention U-Net (BAGAU-Net) that leverages only FLAIR images with a spatially-registered white matter (WM) brain atlas to yield competitive WMH segmentation performance. Specifically, we designed a dual-path segmentation model with two novel connecting mechanisms, namely multi-input attention module (MAM) and attention fusion module (AFM) to fuse the information from two paths for accurate results. Experiments on two publicly available datasets show the effectiveness of the proposed BAGAU-Net. With only FLAIR images and WM brain atlas, BAGAU-Net outperforms the state-of-the-art method with T1-weighted images, paving the way for effective development of WMH segmentation. Availability:https://github.com/Ericzhang1/BAGAU-Net
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge