Lin
Jack
Report for NSF Workshop on AI for Electronic Design Automation
Jan 20, 2026Abstract:This report distills the discussions and recommendations from the NSF Workshop on AI for Electronic Design Automation (EDA), held on December 10, 2024 in Vancouver alongside NeurIPS 2024. Bringing together experts across machine learning and EDA, the workshop examined how AI-spanning large language models (LLMs), graph neural networks (GNNs), reinforcement learning (RL), neurosymbolic methods, etc.-can facilitate EDA and shorten design turnaround. The workshop includes four themes: (1) AI for physical synthesis and design for manufacturing (DFM), discussing challenges in physical manufacturing process and potential AI applications; (2) AI for high-level and logic-level synthesis (HLS/LLS), covering pragma insertion, program transformation, RTL code generation, etc.; (3) AI toolbox for optimization and design, discussing frontier AI developments that could potentially be applied to EDA tasks; and (4) AI for test and verification, including LLM-assisted verification tools, ML-augmented SAT solving, security/reliability challenges, etc. The report recommends NSF to foster AI/EDA collaboration, invest in foundational AI for EDA, develop robust data infrastructures, promote scalable compute infrastructure, and invest in workforce development to democratize hardware design and enable next-generation hardware systems. The workshop information can be found on the website https://ai4eda-workshop.github.io/.
Looking Forward: Challenges and Opportunities in Agentic AI Reliability
Nov 14, 2025Abstract:This chapter presents perspectives for challenges and future development in building reliable AI systems, particularly, agentic AI systems. Several open research problems related to mitigating the risks of cascading failures are discussed. The chapter also sheds lights on research challenges and opportunities in aspects including dynamic environments, inconsistent task execution, unpredictable emergent behaviors, as well as resource-intensive reliability mechanisms. In addition, several research directions along the line of testing and evaluating reliability of agentic AI systems are also discussed.
Fewer Denoising Steps or Cheaper Per-Step Inference: Towards Compute-Optimal Diffusion Model Deployment
Aug 08, 2025



Abstract:Diffusion models have shown remarkable success across generative tasks, yet their high computational demands challenge deployment on resource-limited platforms. This paper investigates a critical question for compute-optimal diffusion model deployment: Under a post-training setting without fine-tuning, is it more effective to reduce the number of denoising steps or to use a cheaper per-step inference? Intuitively, reducing the number of denoising steps increases the variability of the distributions across steps, making the model more sensitive to compression. In contrast, keeping more denoising steps makes the differences smaller, preserving redundancy, and making post-training compression more feasible. To systematically examine this, we propose PostDiff, a training-free framework for accelerating pre-trained diffusion models by reducing redundancy at both the input level and module level in a post-training manner. At the input level, we propose a mixed-resolution denoising scheme based on the insight that reducing generation resolution in early denoising steps can enhance low-frequency components and improve final generation fidelity. At the module level, we employ a hybrid module caching strategy to reuse computations across denoising steps. Extensive experiments and ablation studies demonstrate that (1) PostDiff can significantly improve the fidelity-efficiency trade-off of state-of-the-art diffusion models, and (2) to boost efficiency while maintaining decent generation fidelity, reducing per-step inference cost is often more effective than reducing the number of denoising steps. Our code is available at https://github.com/GATECH-EIC/PostDiff.
Improvement Strategies for Few-Shot Learning in OCT Image Classification of Rare Retinal Diseases
May 26, 2025


Abstract:This paper focuses on using few-shot learning to improve the accuracy of classifying OCT diagnosis images with major and rare classes. We used the GAN-based augmentation strategy as a baseline and introduced several novel methods to further enhance our model. The proposed strategy contains U-GAT-IT for improving the generative part and uses the data balance technique to narrow down the skew of accuracy between all categories. The best model obtained was built with CBAM attention mechanism and fine-tuned InceptionV3, and achieved an overall accuracy of 97.85%, representing a significant improvement over the original baseline.
The Hitchhikers Guide to Production-ready Trustworthy Foundation Model powered Software (FMware)
May 15, 2025
Abstract:Foundation Models (FMs) such as Large Language Models (LLMs) are reshaping the software industry by enabling FMware, systems that integrate these FMs as core components. In this KDD 2025 tutorial, we present a comprehensive exploration of FMware that combines a curated catalogue of challenges with real-world production concerns. We first discuss the state of research and practice in building FMware. We further examine the difficulties in selecting suitable models, aligning high-quality domain-specific data, engineering robust prompts, and orchestrating autonomous agents. We then address the complex journey from impressive demos to production-ready systems by outlining issues in system testing, optimization, deployment, and integration with legacy software. Drawing on our industrial experience and recent research in the area, we provide actionable insights and a technology roadmap for overcoming these challenges. Attendees will gain practical strategies to enable the creation of trustworthy FMware in the evolving technology landscape.
CLIMB: CLustering-based Iterative Data Mixture Bootstrapping for Language Model Pre-training
Apr 17, 2025



Abstract:Pre-training datasets are typically collected from web content and lack inherent domain divisions. For instance, widely used datasets like Common Crawl do not include explicit domain labels, while manually curating labeled datasets such as The Pile is labor-intensive. Consequently, identifying an optimal pre-training data mixture remains a challenging problem, despite its significant benefits for pre-training performance. To address these challenges, we propose CLustering-based Iterative Data Mixture Bootstrapping (CLIMB), an automated framework that discovers, evaluates, and refines data mixtures in a pre-training setting. Specifically, CLIMB embeds and clusters large-scale datasets in a semantic space and then iteratively searches for optimal mixtures using a smaller proxy model and a predictor. When continuously trained on 400B tokens with this mixture, our 1B model exceeds the state-of-the-art Llama-3.2-1B by 2.0%. Moreover, we observe that optimizing for a specific domain (e.g., Social Sciences) yields a 5% improvement over random sampling. Finally, we introduce ClimbLab, a filtered 1.2-trillion-token corpus with 20 clusters as a research playground, and ClimbMix, a compact yet powerful 400-billion-token dataset designed for efficient pre-training that delivers superior performance under an equal token budget. We analyze the final data mixture, elucidating the characteristics of an optimal data mixture. Our data is available at: https://research.nvidia.com/labs/lpr/climb/
Early-Bird Diffusion: Investigating and Leveraging Timestep-Aware Early-Bird Tickets in Diffusion Models for Efficient Training
Apr 13, 2025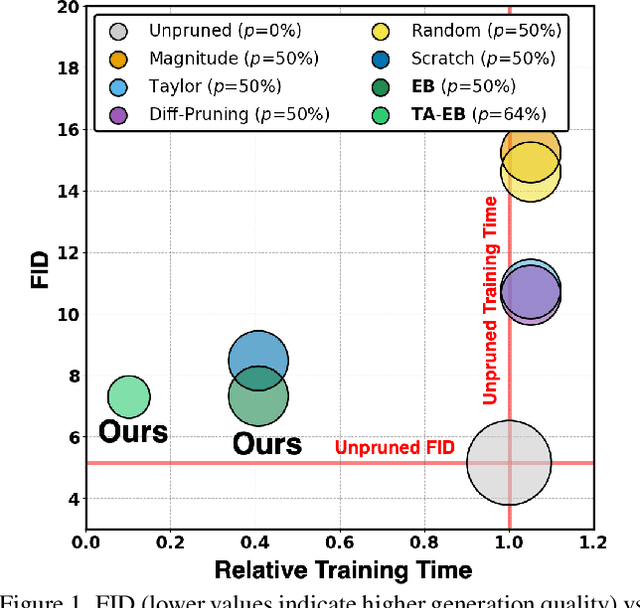
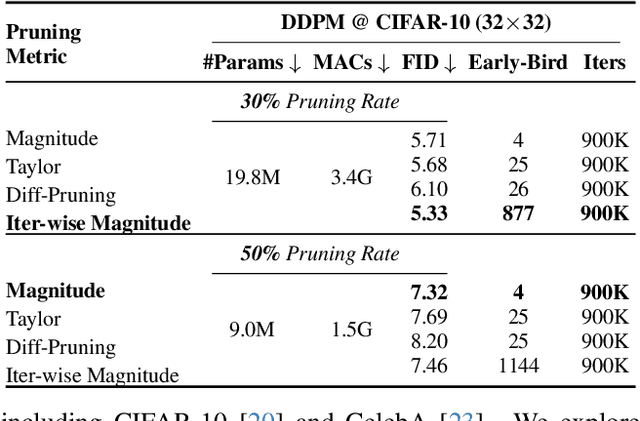
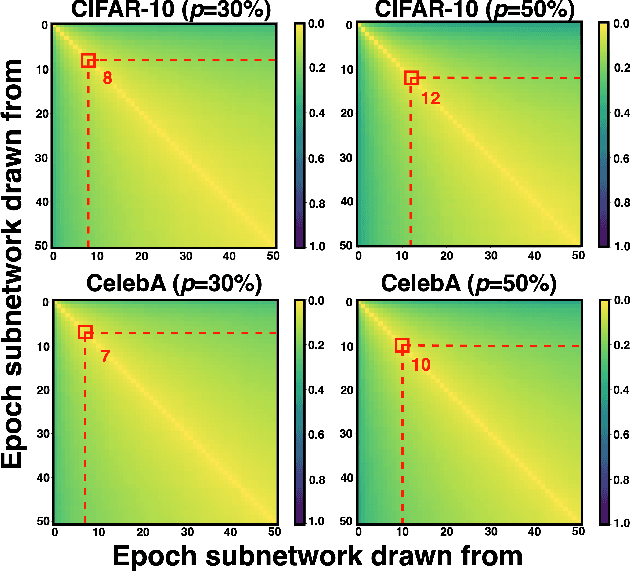
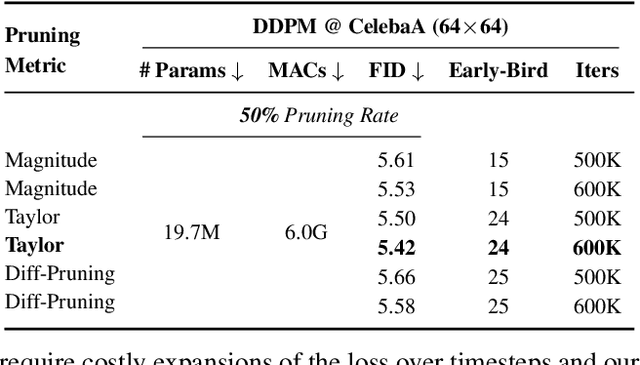
Abstract:Training diffusion models (DMs) requires substantial computational resources due to multiple forward and backward passes across numerous timesteps, motivating research into efficient training techniques. In this paper, we propose EB-Diff-Train, a new efficient DM training approach that is orthogonal to other methods of accelerating DM training, by investigating and leveraging Early-Bird (EB) tickets -- sparse subnetworks that manifest early in the training process and maintain high generation quality. We first investigate the existence of traditional EB tickets in DMs, enabling competitive generation quality without fully training a dense model. Then, we delve into the concept of diffusion-dedicated EB tickets, drawing on insights from varying importance of different timestep regions. These tickets adapt their sparsity levels according to the importance of corresponding timestep regions, allowing for aggressive sparsity during non-critical regions while conserving computational resources for crucial timestep regions. Building on this, we develop an efficient DM training technique that derives timestep-aware EB tickets, trains them in parallel, and combines them during inference for image generation. Extensive experiments validate the existence of both traditional and timestep-aware EB tickets, as well as the effectiveness of our proposed EB-Diff-Train method. This approach can significantly reduce training time both spatially and temporally -- achieving 2.9$\times$ to 5.8$\times$ speedups over training unpruned dense models, and up to 10.3$\times$ faster training compared to standard train-prune-finetune pipelines -- without compromising generative quality. Our code is available at https://github.com/GATECH-EIC/Early-Bird-Diffusion.
Impact of Stain Variation and Color Normalization for Prognostic Predictions in Pathology
Sep 12, 2024



Abstract:In recent years, deep neural networks (DNNs) have demonstrated remarkable performance in pathology applications, potentially even outperforming expert pathologists due to their ability to learn subtle features from large datasets. One complication in preparing digital pathology datasets for DNN tasks is variation in tinctorial qualities. A common way to address this is to perform stain normalization on the images. In this study, we show that a well-trained DNN model trained on one batch of histological slides failed to generalize to another batch prepared at a different time from the same tissue blocks, even when stain normalization methods were applied. This study used sample data from a previously reported DNN that was able to identify patients with early stage non-small cell lung cancer (NSCLC) whose tumors did and did not metastasize, with high accuracy, based on training and then testing of digital images from H&E stained primary tumor tissue sections processed at the same time. In this study we obtained a new series of histologic slides from the adjacent recuts of same tissue blocks processed in the same lab but at a different time. We found that the DNN trained on the either batch of slides/images was unable to generalize and failed to predict progression in the other batch of slides/images (AUC_cross-batch = 0.52 - 0.53 compared to AUC_same-batch = 0.74 - 0.81). The failure to generalize did not improve even when the tinctorial difference correction were made through either traditional color-tuning or stain normalization with the help of a Cycle Generative Adversarial Network (CycleGAN) process. This highlights the need to develop an entirely new way to process and collect consistent microscopy images from histologic slides that can be used to both train and allow for the general application of predictive DNN algorithms.
MG-Verilog: Multi-grained Dataset Towards Enhanced LLM-assisted Verilog Generation
Jul 02, 2024



Abstract:Large Language Models (LLMs) have recently shown promise in streamlining hardware design processes by encapsulating vast amounts of domain-specific data. In addition, they allow users to interact with the design processes through natural language instructions, thus making hardware design more accessible to developers. However, effectively leveraging LLMs in hardware design necessitates providing domain-specific data during inference (e.g., through in-context learning), fine-tuning, or pre-training. Unfortunately, existing publicly available hardware datasets are often limited in size, complexity, or detail, which hinders the effectiveness of LLMs in hardware design tasks. To address this issue, we first propose a set of criteria for creating high-quality hardware datasets that can effectively enhance LLM-assisted hardware design. Based on these criteria, we propose a Multi-Grained-Verilog (MG-Verilog) dataset, which encompasses descriptions at various levels of detail and corresponding code samples. To benefit the broader hardware design community, we have developed an open-source infrastructure that facilitates easy access, integration, and extension of the dataset to meet specific project needs. Furthermore, to fully exploit the potential of the MG-Verilog dataset, which varies in complexity and detail, we introduce a balanced fine-tuning scheme. This scheme serves as a unique use case to leverage the diverse levels of detail provided by the dataset. Extensive experiments demonstrate that the proposed dataset and fine-tuning scheme consistently improve the performance of LLMs in hardware design tasks.
When Linear Attention Meets Autoregressive Decoding: Towards More Effective and Efficient Linearized Large Language Models
Jun 11, 2024Abstract:Autoregressive Large Language Models (LLMs) have achieved impressive performance in language tasks but face two significant bottlenecks: (1) quadratic complexity in the attention module as the number of tokens increases, and (2) limited efficiency due to the sequential processing nature of autoregressive LLMs during generation. While linear attention and speculative decoding offer potential solutions, their applicability and synergistic potential for enhancing autoregressive LLMs remain uncertain. We conduct the first comprehensive study on the efficacy of existing linear attention methods for autoregressive LLMs, integrating them with speculative decoding. We introduce an augmentation technique for linear attention that ensures compatibility with speculative decoding, enabling more efficient training and serving of LLMs. Extensive experiments and ablation studies involving seven existing linear attention models and five encoder/decoder-based LLMs consistently validate the effectiveness of our augmented linearized LLMs. Notably, our approach achieves up to a 6.67 reduction in perplexity on the LLaMA model and up to a 2$\times$ speedup during generation compared to prior linear attention methods. Codes and models are available at https://github.com/GATECH-EIC/Linearized-LLM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge