Guoqiang Han
BroadCAM: Outcome-agnostic Class Activation Mapping for Small-scale Weakly Supervised Applications
Sep 07, 2023



Abstract:Class activation mapping~(CAM), a visualization technique for interpreting deep learning models, is now commonly used for weakly supervised semantic segmentation~(WSSS) and object localization~(WSOL). It is the weighted aggregation of the feature maps by activating the high class-relevance ones. Current CAM methods achieve it relying on the training outcomes, such as predicted scores~(forward information), gradients~(backward information), etc. However, when with small-scale data, unstable training may lead to less effective model outcomes and generate unreliable weights, finally resulting in incorrect activation and noisy CAM seeds. In this paper, we propose an outcome-agnostic CAM approach, called BroadCAM, for small-scale weakly supervised applications. Since broad learning system (BLS) is independent to the model learning, BroadCAM can avoid the weights being affected by the unreliable model outcomes when with small-scale data. By evaluating BroadCAM on VOC2012 (natural images) and BCSS-WSSS (medical images) for WSSS and OpenImages30k for WSOL, BroadCAM demonstrates superior performance than existing CAM methods with small-scale data (less than 5\%) in different CNN architectures. It also achieves SOTA performance with large-scale training data. Extensive qualitative comparisons are conducted to demonstrate how BroadCAM activates the high class-relevance feature maps and generates reliable CAMs when with small-scale training data.
Rethinking Mitosis Detection: Towards Diverse Data and Feature Representation
Jul 12, 2023Abstract:Mitosis detection is one of the fundamental tasks in computational pathology, which is extremely challenging due to the heterogeneity of mitotic cell. Most of the current studies solve the heterogeneity in the technical aspect by increasing the model complexity. However, lacking consideration of the biological knowledge and the complex model design may lead to the overfitting problem while limited the generalizability of the detection model. In this paper, we systematically study the morphological appearances in different mitotic phases as well as the ambiguous non-mitotic cells and identify that balancing the data and feature diversity can achieve better generalizability. Based on this observation, we propose a novel generalizable framework (MitDet) for mitosis detection. The data diversity is considered by the proposed diversity-guided sample balancing (DGSB). And the feature diversity is preserved by inter- and intra- class feature diversity-preserved module (InCDP). Stain enhancement (SE) module is introduced to enhance the domain-relevant diversity of both data and features simultaneously. Extensive experiments have demonstrated that our proposed model outperforms all the SOTA approaches in several popular mitosis detection datasets in both internal and external test sets using minimal annotation efforts with point annotations only. Comprehensive ablation studies have also proven the effectiveness of the rethinking of data and feature diversity balancing. By analyzing the results quantitatively and qualitatively, we believe that our proposed model not only achieves SOTA performance but also might inspire the future studies in new perspectives. Source code is at https://github.com/Onehour0108/MitDet.
FedDBL: Communication and Data Efficient Federated Deep-Broad Learning for Histopathological Tissue Classification
Feb 24, 2023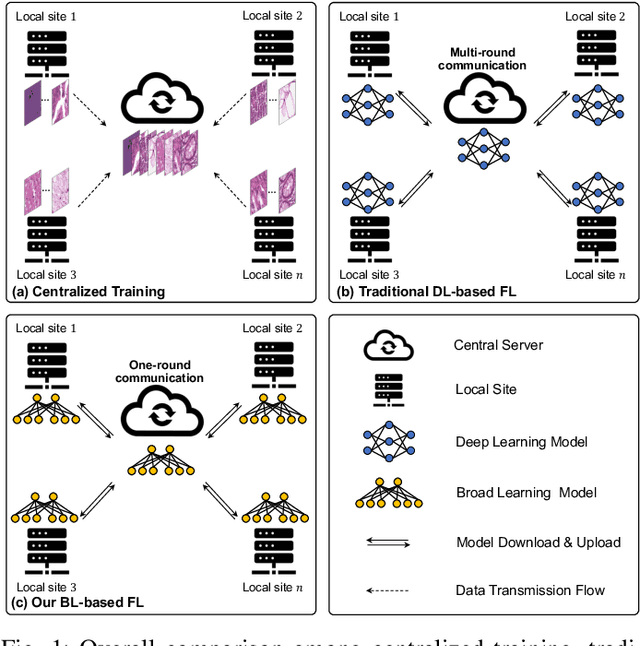
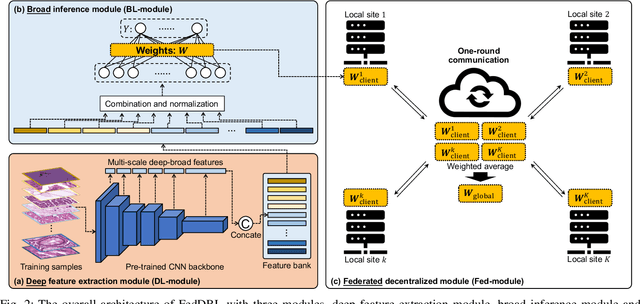
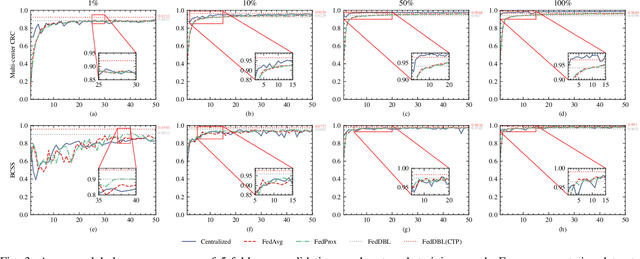
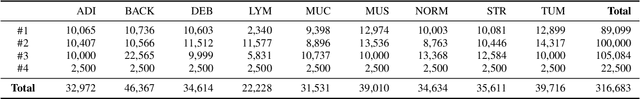
Abstract:Histopathological tissue classification is a fundamental task in computational pathology. Deep learning-based models have achieved superior performance but centralized training with data centralization suffers from the privacy leakage problem. Federated learning (FL) can safeguard privacy by keeping training samples locally, but existing FL-based frameworks require a large number of well-annotated training samples and numerous rounds of communication which hinder their practicability in the real-world clinical scenario. In this paper, we propose a universal and lightweight federated learning framework, named Federated Deep-Broad Learning (FedDBL), to achieve superior classification performance with limited training samples and only one-round communication. By simply associating a pre-trained deep learning feature extractor, a fast and lightweight broad learning inference system and a classical federated aggregation approach, FedDBL can dramatically reduce data dependency and improve communication efficiency. Five-fold cross-validation demonstrates that FedDBL greatly outperforms the competitors with only one-round communication and limited training samples, while it even achieves comparable performance with the ones under multiple-round communications. Furthermore, due to the lightweight design and one-round communication, FedDBL reduces the communication burden from 4.6GB to only 276.5KB per client using the ResNet-50 backbone at 50-round training. Since no data or deep model sharing across different clients, the privacy issue is well-solved and the model security is guaranteed with no model inversion attack risk. Code is available at https://github.com/tianpeng-deng/FedDBL.
CKD-TransBTS: Clinical Knowledge-Driven Hybrid Transformer with Modality-Correlated Cross-Attention for Brain Tumor Segmentation
Jul 15, 2022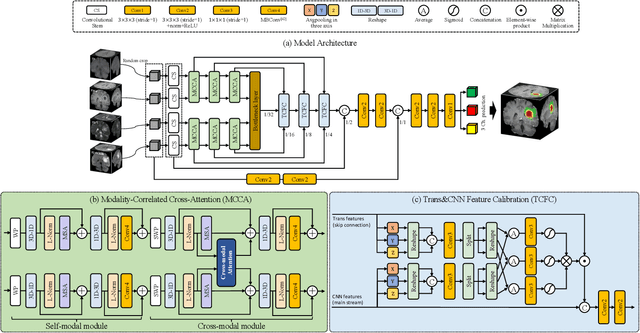
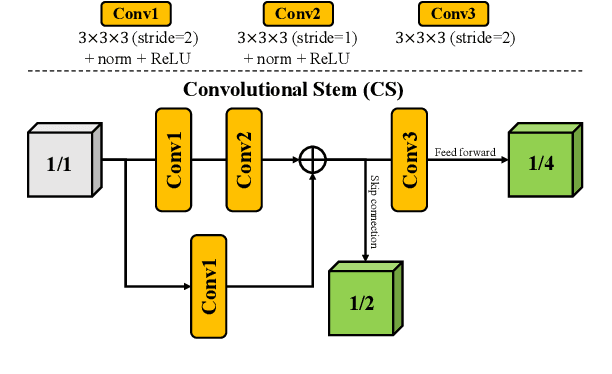
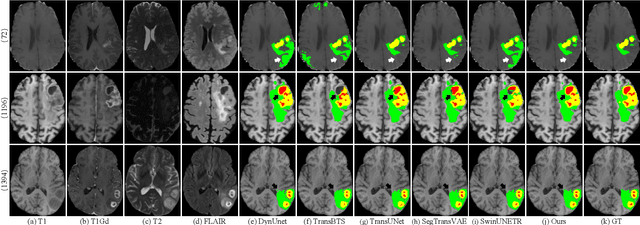
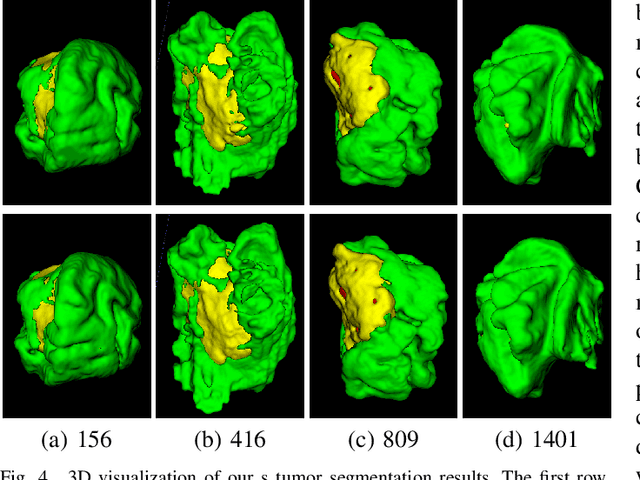
Abstract:Brain tumor segmentation (BTS) in magnetic resonance image (MRI) is crucial for brain tumor diagnosis, cancer management and research purposes. With the great success of the ten-year BraTS challenges as well as the advances of CNN and Transformer algorithms, a lot of outstanding BTS models have been proposed to tackle the difficulties of BTS in different technical aspects. However, existing studies hardly consider how to fuse the multi-modality images in a reasonable manner. In this paper, we leverage the clinical knowledge of how radiologists diagnose brain tumors from multiple MRI modalities and propose a clinical knowledge-driven brain tumor segmentation model, called CKD-TransBTS. Instead of directly concatenating all the modalities, we re-organize the input modalities by separating them into two groups according to the imaging principle of MRI. A dual-branch hybrid encoder with the proposed modality-correlated cross-attention block (MCCA) is designed to extract the multi-modality image features. The proposed model inherits the strengths from both Transformer and CNN with the local feature representation ability for precise lesion boundaries and long-range feature extraction for 3D volumetric images. To bridge the gap between Transformer and CNN features, we propose a Trans&CNN Feature Calibration block (TCFC) in the decoder. We compare the proposed model with five CNN-based models and six transformer-based models on the BraTS 2021 challenge dataset. Extensive experiments demonstrate that the proposed model achieves state-of-the-art brain tumor segmentation performance compared with all the competitors.
WSSS4LUAD: Grand Challenge on Weakly-supervised Tissue Semantic Segmentation for Lung Adenocarcinoma
Apr 14, 2022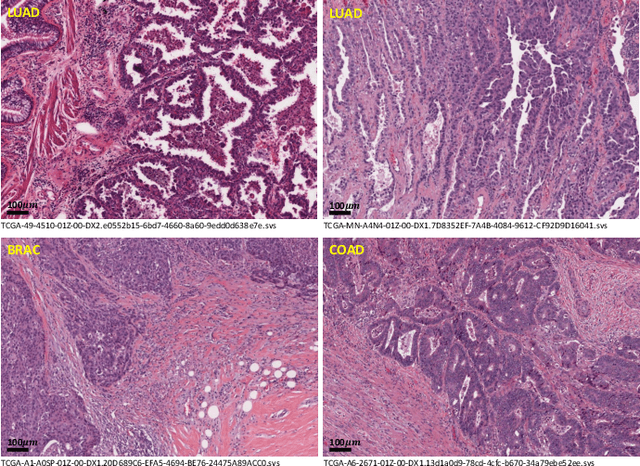
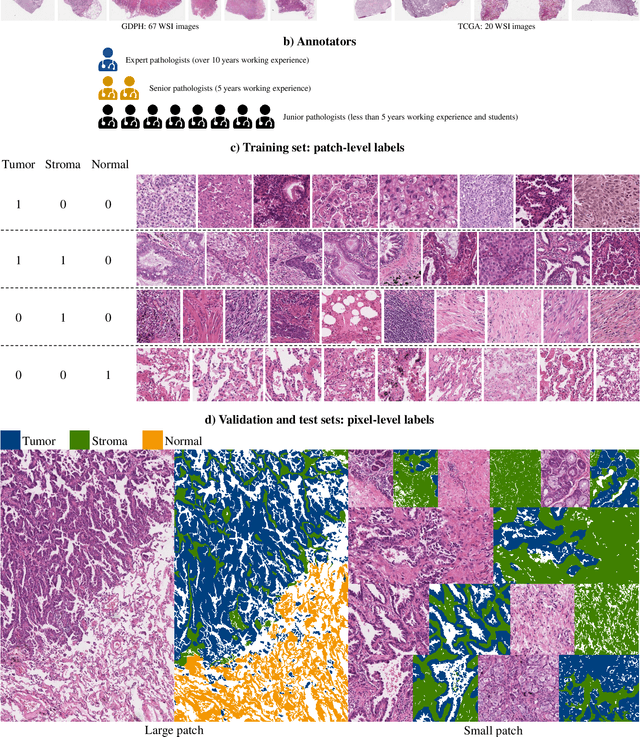
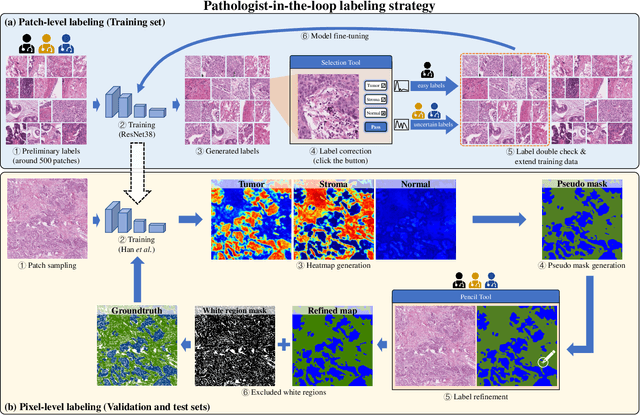
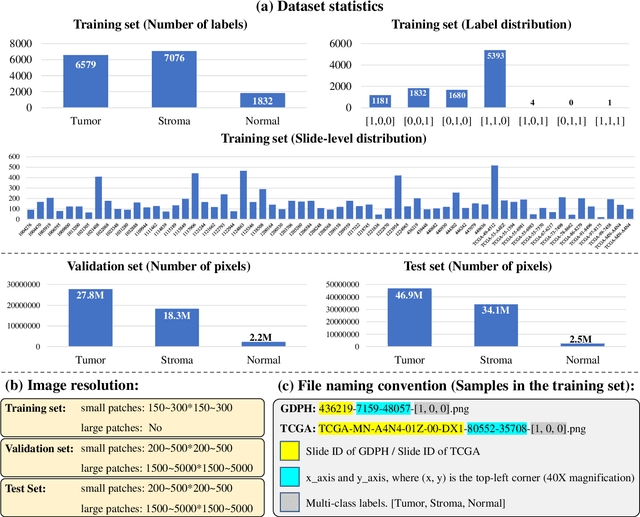
Abstract:Lung cancer is the leading cause of cancer death worldwide, and adenocarcinoma (LUAD) is the most common subtype. Exploiting the potential value of the histopathology images can promote precision medicine in oncology. Tissue segmentation is the basic upstream task of histopathology image analysis. Existing deep learning models have achieved superior segmentation performance but require sufficient pixel-level annotations, which is time-consuming and expensive. To enrich the label resources of LUAD and to alleviate the annotation efforts, we organize this challenge WSSS4LUAD to call for the outstanding weakly-supervised semantic segmentation (WSSS) techniques for histopathology images of LUAD. Participants have to design the algorithm to segment tumor epithelial, tumor-associated stroma and normal tissue with only patch-level labels. This challenge includes 10,091 patch-level annotations (the training set) and over 130 million labeled pixels (the validation and test sets), from 87 WSIs (67 from GDPH, 20 from TCGA). All the labels were generated by a pathologist-in-the-loop pipeline with the help of AI models and checked by the label review board. Among 532 registrations, 28 teams submitted the results in the test phase with over 1,000 submissions. Finally, the first place team achieved mIoU of 0.8413 (tumor: 0.8389, stroma: 0.7931, normal: 0.8919). According to the technical reports of the top-tier teams, CAM is still the most popular approach in WSSS. Cutmix data augmentation has been widely adopted to generate more reliable samples. With the success of this challenge, we believe that WSSS approaches with patch-level annotations can be a complement to the traditional pixel annotations while reducing the annotation efforts. The entire dataset has been released to encourage more researches on computational pathology in LUAD and more novel WSSS techniques.
PDBL: Improving Histopathological Tissue Classification with Plug-and-Play Pyramidal Deep-Broad Learning
Nov 04, 2021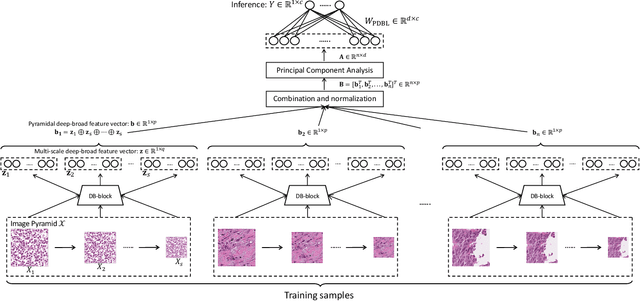
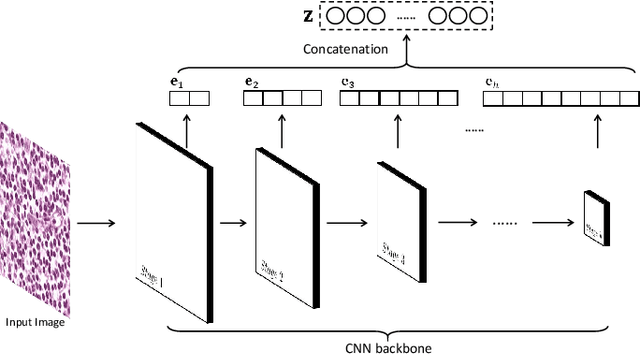

Abstract:Histopathological tissue classification is a fundamental task in pathomics cancer research. Precisely differentiating different tissue types is a benefit for the downstream researches, like cancer diagnosis, prognosis and etc. Existing works mostly leverage the popular classification backbones in computer vision to achieve histopathological tissue classification. In this paper, we proposed a super lightweight plug-and-play module, named Pyramidal Deep-Broad Learning (PDBL), for any well-trained classification backbone to further improve the classification performance without a re-training burden. We mimic how pathologists observe pathology slides in different magnifications and construct an image pyramid for the input image in order to obtain the pyramidal contextual information. For each level in the pyramid, we extract the multi-scale deep-broad features by our proposed Deep-Broad block (DB-block). We equipped PDBL in three popular classification backbones, ShuffLeNetV2, EfficientNetb0, and ResNet50 to evaluate the effectiveness and efficiency of our proposed module on two datasets (Kather Multiclass Dataset and the LC25000 Dataset). Experimental results demonstrate the proposed PDBL can steadily improve the tissue-level classification performance for any CNN backbones, especially for the lightweight models when given a small among of training samples (less than 10%), which greatly saves the computational time and annotation efforts.
Multi-Layer Pseudo-Supervision for Histopathology Tissue Semantic Segmentation using Patch-level Classification Labels
Oct 14, 2021

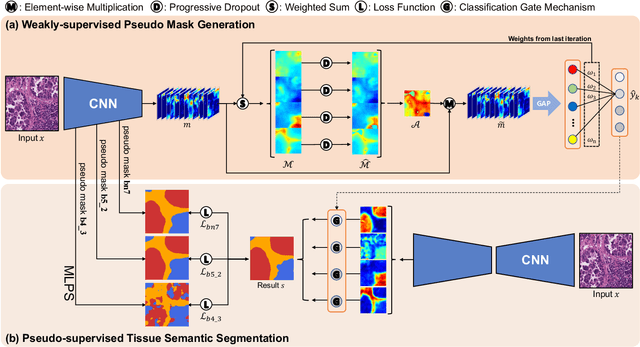
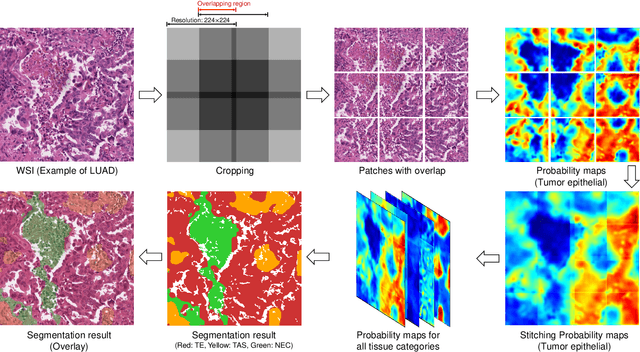
Abstract:Tissue-level semantic segmentation is a vital step in computational pathology. Fully-supervised models have already achieved outstanding performance with dense pixel-level annotations. However, drawing such labels on the giga-pixel whole slide images is extremely expensive and time-consuming. In this paper, we use only patch-level classification labels to achieve tissue semantic segmentation on histopathology images, finally reducing the annotation efforts. We proposed a two-step model including a classification and a segmentation phases. In the classification phase, we proposed a CAM-based model to generate pseudo masks by patch-level labels. In the segmentation phase, we achieved tissue semantic segmentation by our proposed Multi-Layer Pseudo-Supervision. Several technical novelties have been proposed to reduce the information gap between pixel-level and patch-level annotations. As a part of this paper, we introduced a new weakly-supervised semantic segmentation (WSSS) dataset for lung adenocarcinoma (LUAD-HistoSeg). We conducted several experiments to evaluate our proposed model on two datasets. Our proposed model outperforms two state-of-the-art WSSS approaches. Note that we can achieve comparable quantitative and qualitative results with the fully-supervised model, with only around a 2\% gap for MIoU and FwIoU. By comparing with manual labeling, our model can greatly save the annotation time from hours to minutes. The source code is available at: \url{https://github.com/ChuHan89/WSSS-Tissue}.
Unifying Global-Local Representations in Salient Object Detection with Transformer
Aug 05, 2021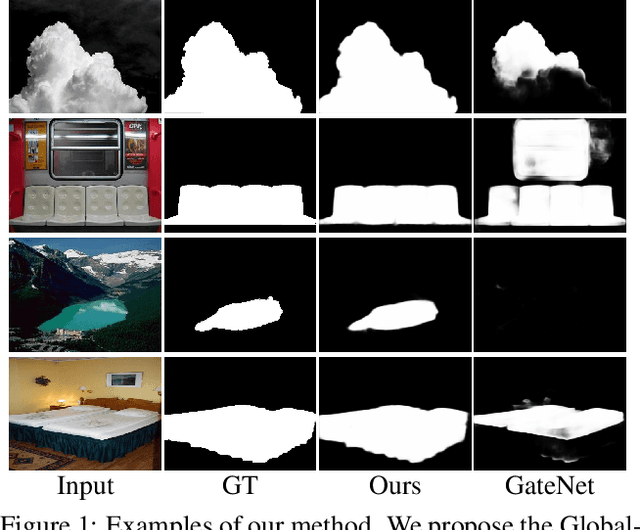
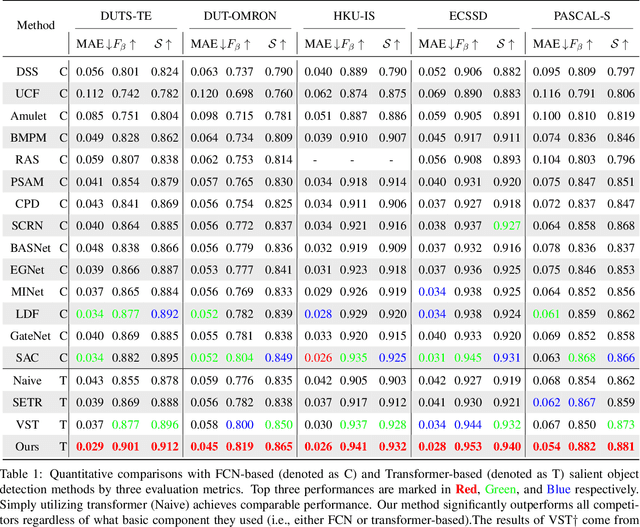
Abstract:The fully convolutional network (FCN) has dominated salient object detection for a long period. However, the locality of CNN requires the model deep enough to have a global receptive field and such a deep model always leads to the loss of local details. In this paper, we introduce a new attention-based encoder, vision transformer, into salient object detection to ensure the globalization of the representations from shallow to deep layers. With the global view in very shallow layers, the transformer encoder preserves more local representations to recover the spatial details in final saliency maps. Besides, as each layer can capture a global view of its previous layer, adjacent layers can implicitly maximize the representation differences and minimize the redundant features, making that every output feature of transformer layers contributes uniquely for final prediction. To decode features from the transformer, we propose a simple yet effective deeply-transformed decoder. The decoder densely decodes and upsamples the transformer features, generating the final saliency map with less noise injection. Experimental results demonstrate that our method significantly outperforms other FCN-based and transformer-based methods in five benchmarks by a large margin, with an average of 12.17% improvement in terms of Mean Absolute Error (MAE). Code will be available at https://github.com/OliverRensu/GLSTR.
TENet: Triple Excitation Network for Video Salient Object Detection
Aug 30, 2020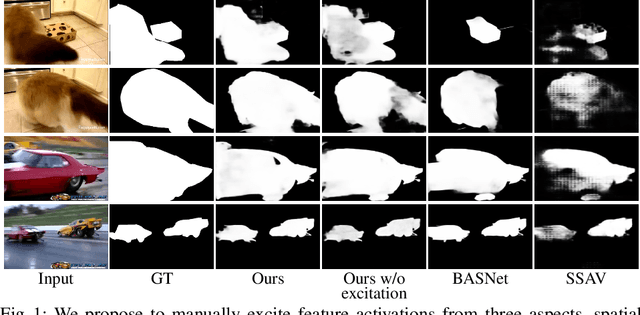
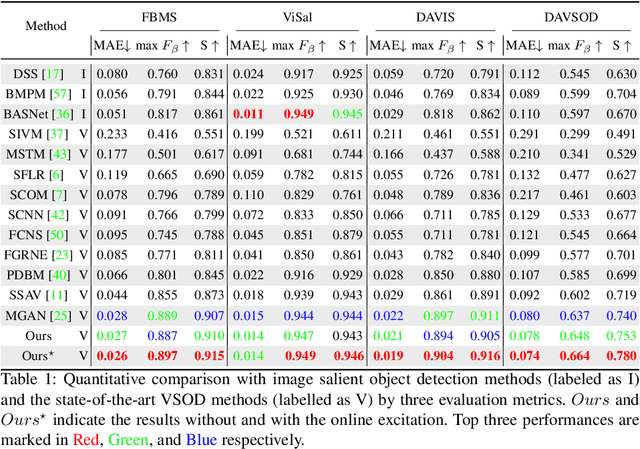
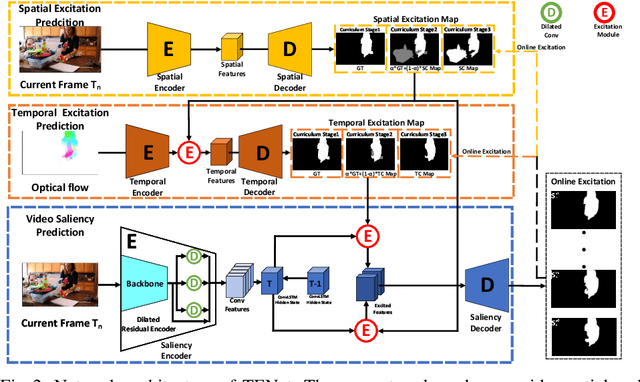
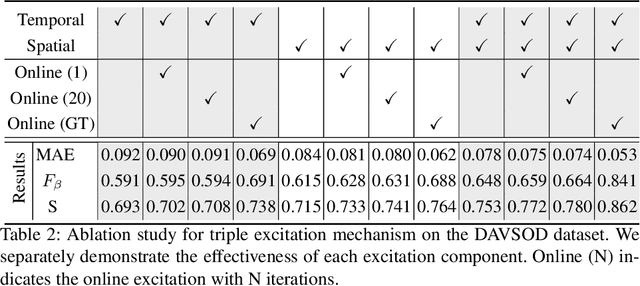
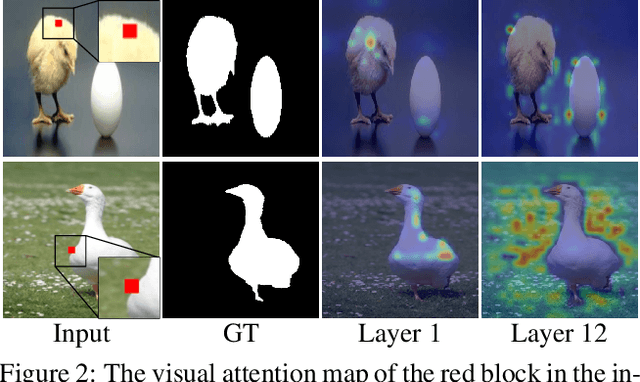
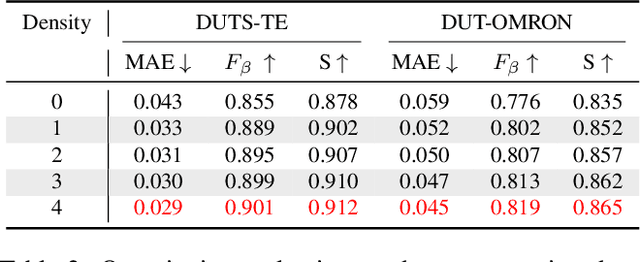
Abstract:In this paper, we propose a simple yet effective approach, named Triple Excitation Network, to reinforce the training of video salient object detection (VSOD) from three aspects, spatial, temporal, and online excitations. These excitation mechanisms are designed following the spirit of curriculum learning and aim to reduce learning ambiguities at the beginning of training by selectively exciting feature activations using ground truth. Then we gradually reduce the weight of ground truth excitations by a curriculum rate and replace it by a curriculum complementary map for better and faster convergence. In particular, the spatial excitation strengthens feature activations for clear object boundaries, while the temporal excitation imposes motions to emphasize spatio-temporal salient regions. Spatial and temporal excitations can combat the saliency shifting problem and conflict between spatial and temporal features of VSOD. Furthermore, our semi-curriculum learning design enables the first online refinement strategy for VSOD, which allows exciting and boosting saliency responses during testing without re-training. The proposed triple excitations can easily plug in different VSOD methods. Extensive experiments show the effectiveness of all three excitation methods and the proposed method outperforms state-of-the-art image and video salient object detection methods.
Context-aware and Scale-insensitive Temporal Repetition Counting
May 18, 2020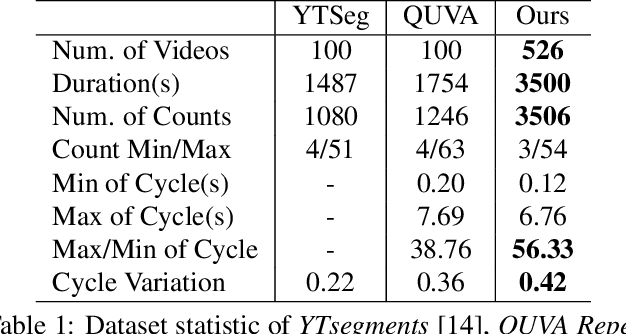
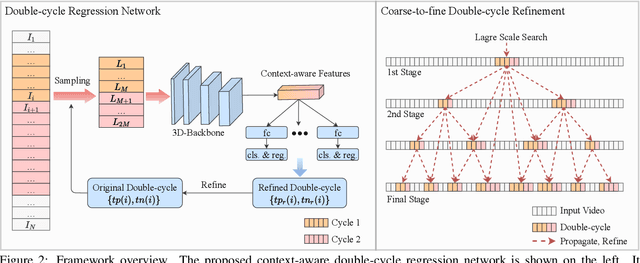
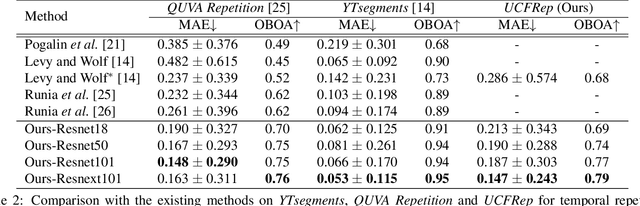

Abstract:Temporal repetition counting aims to estimate the number of cycles of a given repetitive action. Existing deep learning methods assume repetitive actions are performed in a fixed time-scale, which is invalid for the complex repetitive actions in real life. In this paper, we tailor a context-aware and scale-insensitive framework, to tackle the challenges in repetition counting caused by the unknown and diverse cycle-lengths. Our approach combines two key insights: (1) Cycle lengths from different actions are unpredictable that require large-scale searching, but, once a coarse cycle length is determined, the variety between repetitions can be overcome by regression. (2) Determining the cycle length cannot only rely on a short fragment of video but a contextual understanding. The first point is implemented by a coarse-to-fine cycle refinement method. It avoids the heavy computation of exhaustively searching all the cycle lengths in the video, and, instead, it propagates the coarse prediction for further refinement in a hierarchical manner. We secondly propose a bidirectional cycle length estimation method for a context-aware prediction. It is a regression network that takes two consecutive coarse cycles as input, and predicts the locations of the previous and next repetitive cycles. To benefit the training and evaluation of temporal repetition counting area, we construct a new and largest benchmark, which contains 526 videos with diverse repetitive actions. Extensive experiments show that the proposed network trained on a single dataset outperforms state-of-the-art methods on several benchmarks, indicating that the proposed framework is general enough to capture repetition patterns across domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge