Binghong Chen
Multi-View Learning with Context-Guided Receptance for Image Denoising
May 05, 2025Abstract:Image denoising is essential in low-level vision applications such as photography and automated driving. Existing methods struggle with distinguishing complex noise patterns in real-world scenes and consume significant computational resources due to reliance on Transformer-based models. In this work, the Context-guided Receptance Weighted Key-Value (\M) model is proposed, combining enhanced multi-view feature integration with efficient sequence modeling. Our approach introduces the Context-guided Token Shift (CTS) paradigm, which effectively captures local spatial dependencies and enhance the model's ability to model real-world noise distributions. Additionally, the Frequency Mix (FMix) module extracting frequency-domain features is designed to isolate noise in high-frequency spectra, and is integrated with spatial representations through a multi-view learning process. To improve computational efficiency, the Bidirectional WKV (BiWKV) mechanism is adopted, enabling full pixel-sequence interaction with linear complexity while overcoming the causal selection constraints. The model is validated on multiple real-world image denoising datasets, outperforming the existing state-of-the-art methods quantitatively and reducing inference time up to 40\%. Qualitative results further demonstrate the ability of our model to restore fine details in various scenes.
PolyGET: Accelerating Polymer Simulations by Accurate and Generalizable Forcefield with Equivariant Transformer
Sep 01, 2023



Abstract:Polymer simulation with both accuracy and efficiency is a challenging task. Machine learning (ML) forcefields have been developed to achieve both the accuracy of ab initio methods and the efficiency of empirical force fields. However, existing ML force fields are usually limited to single-molecule settings, and their simulations are not robust enough. In this paper, we present PolyGET, a new framework for Polymer Forcefields with Generalizable Equivariant Transformers. PolyGET is designed to capture complex quantum interactions between atoms and generalize across various polymer families, using a deep learning model called Equivariant Transformers. We propose a new training paradigm that focuses exclusively on optimizing forces, which is different from existing methods that jointly optimize forces and energy. This simple force-centric objective function avoids competing objectives between energy and forces, thereby allowing for learning a unified forcefield ML model over different polymer families. We evaluated PolyGET on a large-scale dataset of 24 distinct polymer types and demonstrated state-of-the-art performance in force accuracy and robust MD simulations. Furthermore, PolyGET can simulate large polymers with high fidelity to the reference ab initio DFT method while being able to generalize to unseen polymers.
May the Force be with You: Unified Force-Centric Pre-Training for 3D Molecular Conformations
Aug 24, 2023Abstract:Recent works have shown the promise of learning pre-trained models for 3D molecular representation. However, existing pre-training models focus predominantly on equilibrium data and largely overlook off-equilibrium conformations. It is challenging to extend these methods to off-equilibrium data because their training objective relies on assumptions of conformations being the local energy minima. We address this gap by proposing a force-centric pretraining model for 3D molecular conformations covering both equilibrium and off-equilibrium data. For off-equilibrium data, our model learns directly from their atomic forces. For equilibrium data, we introduce zero-force regularization and forced-based denoising techniques to approximate near-equilibrium forces. We obtain a unified pre-trained model for 3D molecular representation with over 15 million diverse conformations. Experiments show that, with our pre-training objective, we increase forces accuracy by around 3 times compared to the un-pre-trained Equivariant Transformer model. By incorporating regularizations on equilibrium data, we solved the problem of unstable MD simulations in vanilla Equivariant Transformers, achieving state-of-the-art simulation performance with 2.45 times faster inference time than NequIP. As a powerful molecular encoder, our pre-trained model achieves on-par performance with state-of-the-art property prediction tasks.
Learning to Improve Code Efficiency
Aug 09, 2022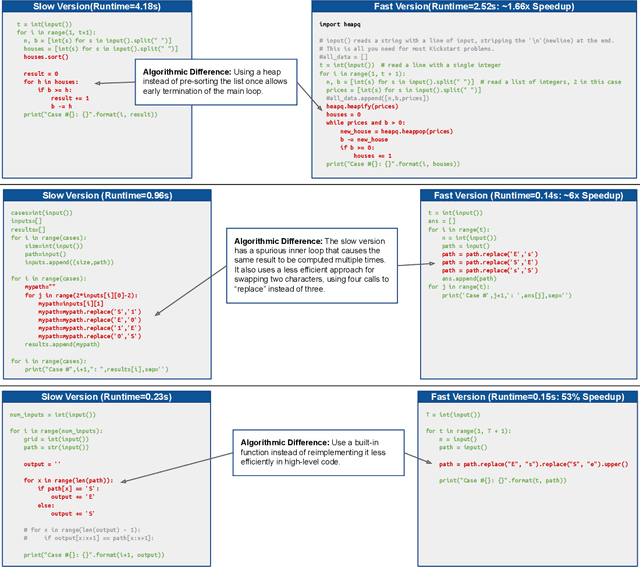
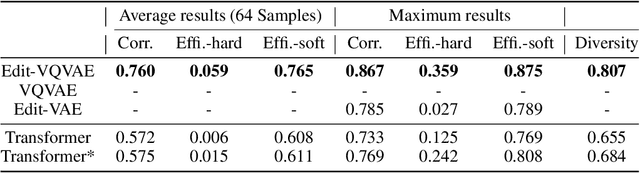
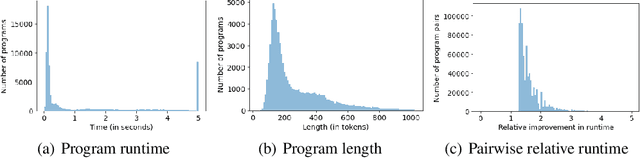
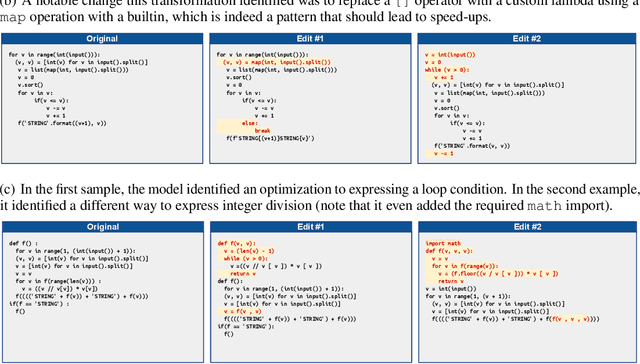
Abstract:Improvements in the performance of computing systems, driven by Moore's Law, have transformed society. As such hardware-driven gains slow down, it becomes even more important for software developers to focus on performance and efficiency during development. While several studies have demonstrated the potential from such improved code efficiency (e.g., 2x better generational improvements compared to hardware), unlocking these gains in practice has been challenging. Reasoning about algorithmic complexity and the interaction of coding patterns on hardware can be challenging for the average programmer, especially when combined with pragmatic constraints around development velocity and multi-person development. This paper seeks to address this problem. We analyze a large competitive programming dataset from the Google Code Jam competition and find that efficient code is indeed rare, with a 2x runtime difference between the median and the 90th percentile of solutions. We propose using machine learning to automatically provide prescriptive feedback in the form of hints, to guide programmers towards writing high-performance code. To automatically learn these hints from the dataset, we propose a novel discrete variational auto-encoder, where each discrete latent variable represents a different learned category of code-edit that increases performance. We show that this method represents the multi-modal space of code efficiency edits better than a sequence-to-sequence baseline and generates a distribution of more efficient solutions.
Learning Temporal Rules from Noisy Timeseries Data
Feb 11, 2022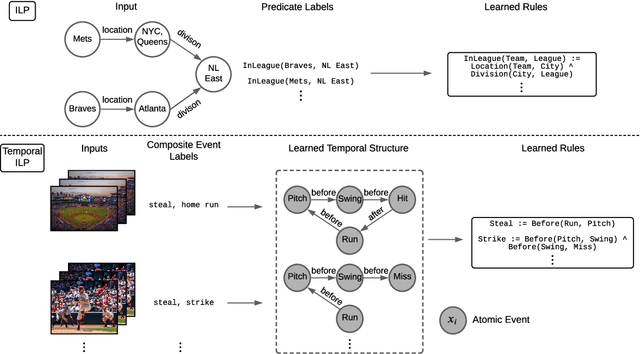
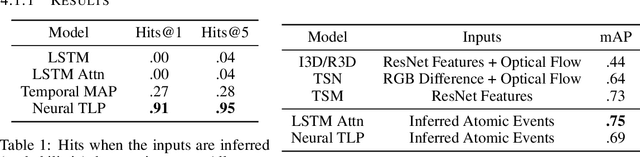
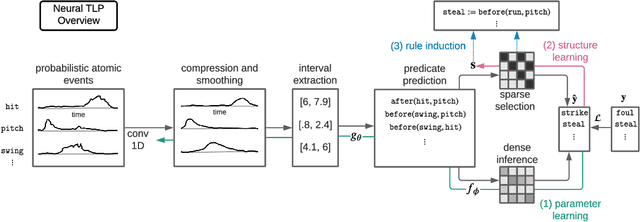
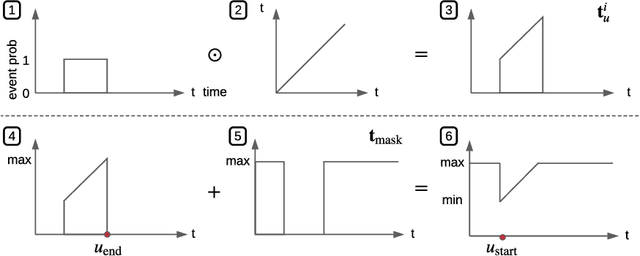
Abstract:Events across a timeline are a common data representation, seen in different temporal modalities. Individual atomic events can occur in a certain temporal ordering to compose higher level composite events. Examples of a composite event are a patient's medical symptom or a baseball player hitting a home run, caused distinct temporal orderings of patient vitals and player movements respectively. Such salient composite events are provided as labels in temporal datasets and most works optimize models to predict these composite event labels directly. We focus on uncovering the underlying atomic events and their relations that lead to the composite events within a noisy temporal data setting. We propose Neural Temporal Logic Programming (Neural TLP) which first learns implicit temporal relations between atomic events and then lifts logic rules for composite events, given only the composite events labels for supervision. This is done through efficiently searching through the combinatorial space of all temporal logic rules in an end-to-end differentiable manner. We evaluate our method on video and healthcare datasets where it outperforms the baseline methods for rule discovery.
ProTo: Program-Guided Transformer for Program-Guided Tasks
Oct 16, 2021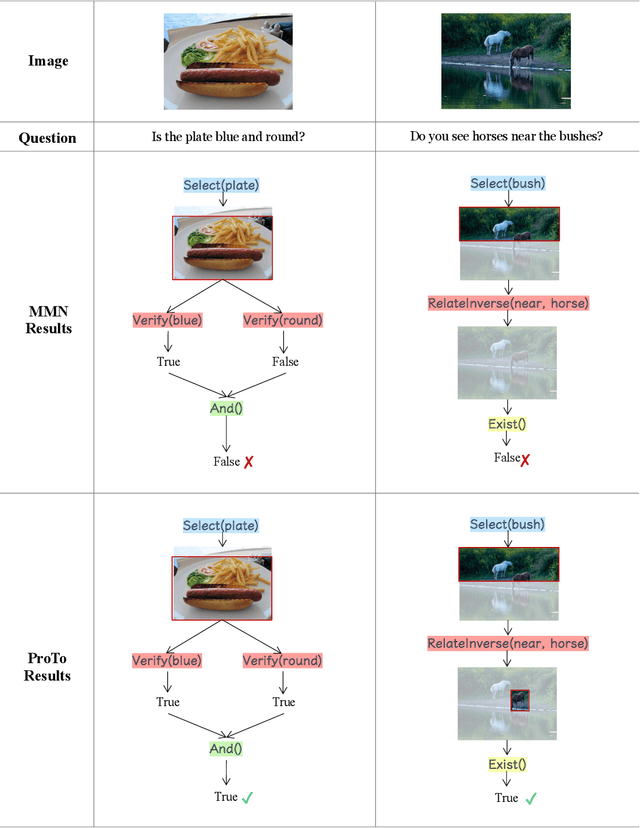
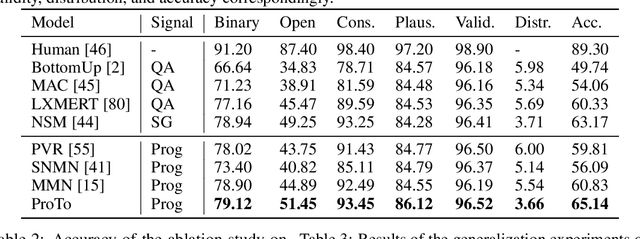
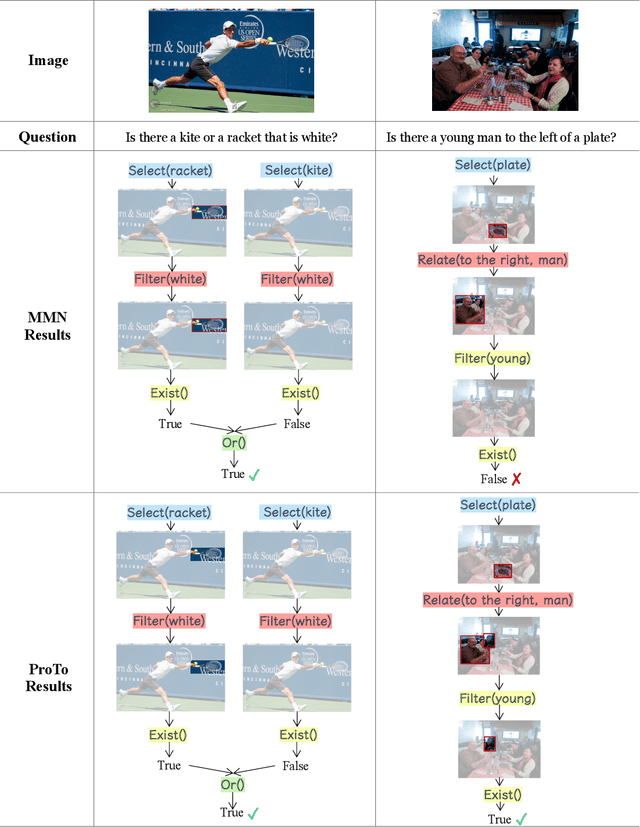
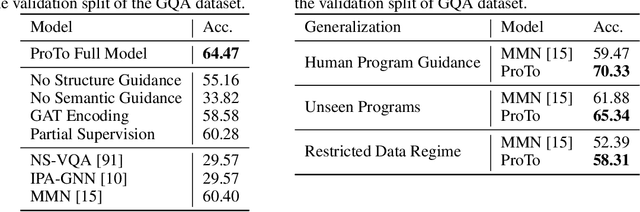
Abstract:Programs, consisting of semantic and structural information, play an important role in the communication between humans and agents. Towards learning general program executors to unify perception, reasoning, and decision making, we formulate program-guided tasks which require learning to execute a given program on the observed task specification. Furthermore, we propose the Program-guided Transformer (ProTo), which integrates both semantic and structural guidance of a program by leveraging cross-attention and masked self-attention to pass messages between the specification and routines in the program. ProTo executes a program in a learned latent space and enjoys stronger representation ability than previous neural-symbolic approaches. We demonstrate that ProTo significantly outperforms the previous state-of-the-art methods on GQA visual reasoning and 2D Minecraft policy learning datasets. Additionally, ProTo demonstrates better generalization to unseen, complex, and human-written programs.
Graph Contrastive Pre-training for Effective Theorem Reasoning
Aug 24, 2021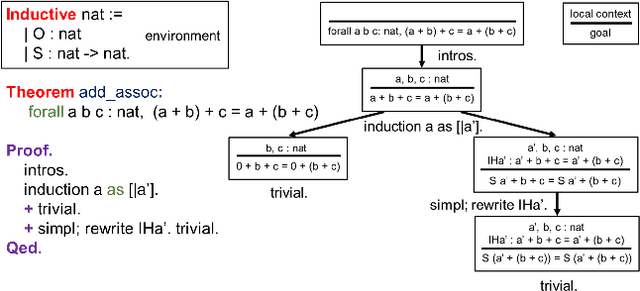
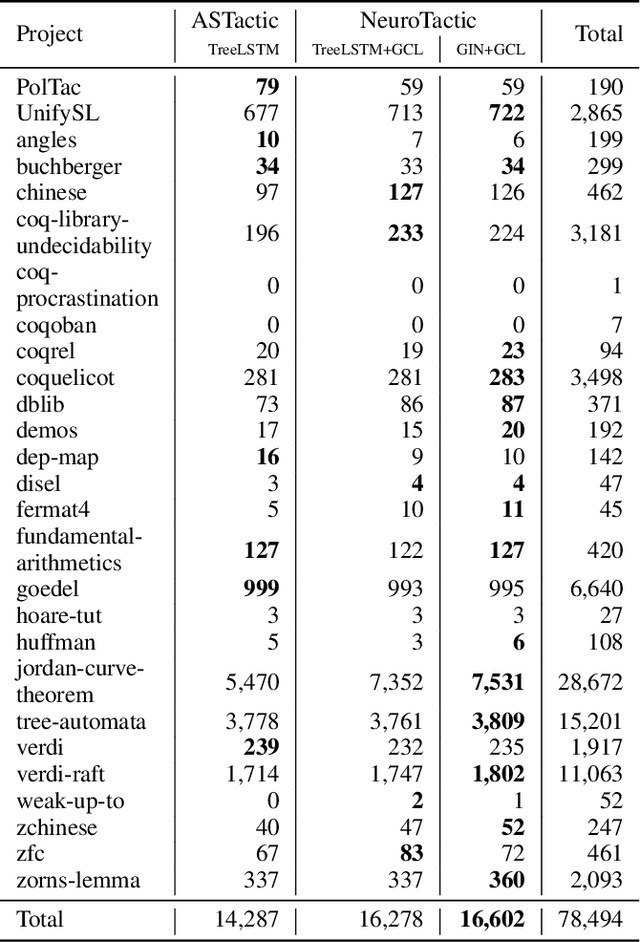
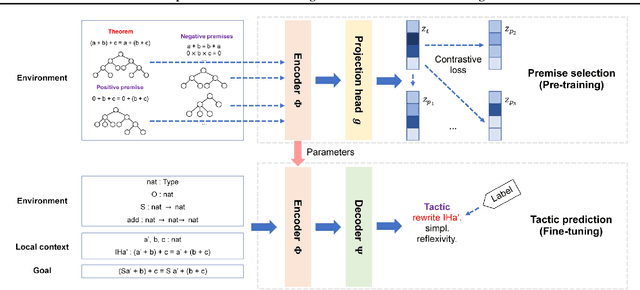
Abstract:Interactive theorem proving is a challenging and tedious process, which requires non-trivial expertise and detailed low-level instructions (or tactics) from human experts. Tactic prediction is a natural way to automate this process. Existing methods show promising results on tactic prediction by learning a deep neural network (DNN) based model from proofs written by human experts. In this paper, we propose NeuroTactic, a novel extension with a special focus on improving the representation learning for theorem proving. NeuroTactic leverages graph neural networks (GNNs) to represent the theorems and premises, and applies graph contrastive learning for pre-training. We demonstrate that the representation learning of theorems is essential to predict tactics. Compared with other methods, NeuroTactic achieves state-of-the-art performance on the CoqGym dataset.
BioNavi-NP: Biosynthesis Navigator for Natural Products
May 26, 2021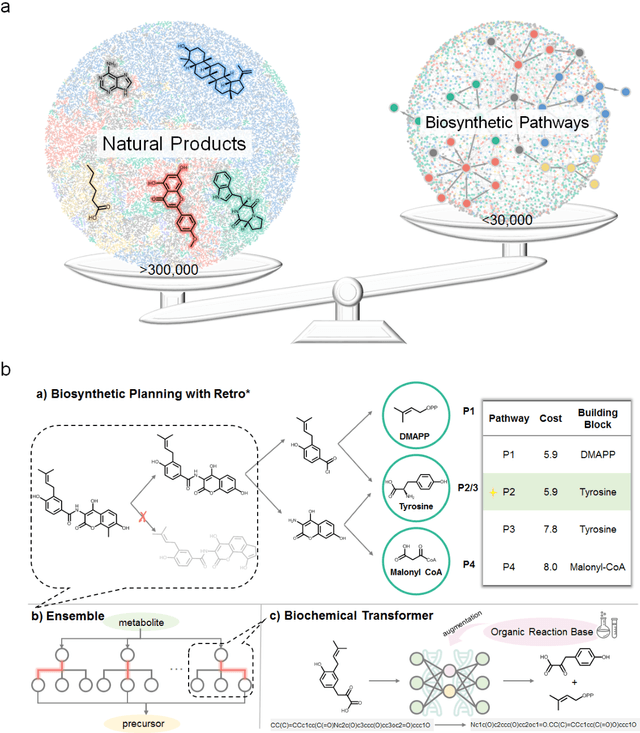
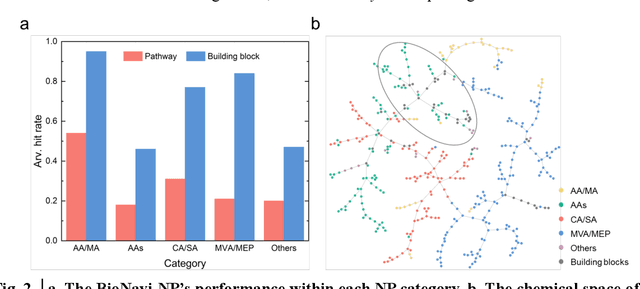
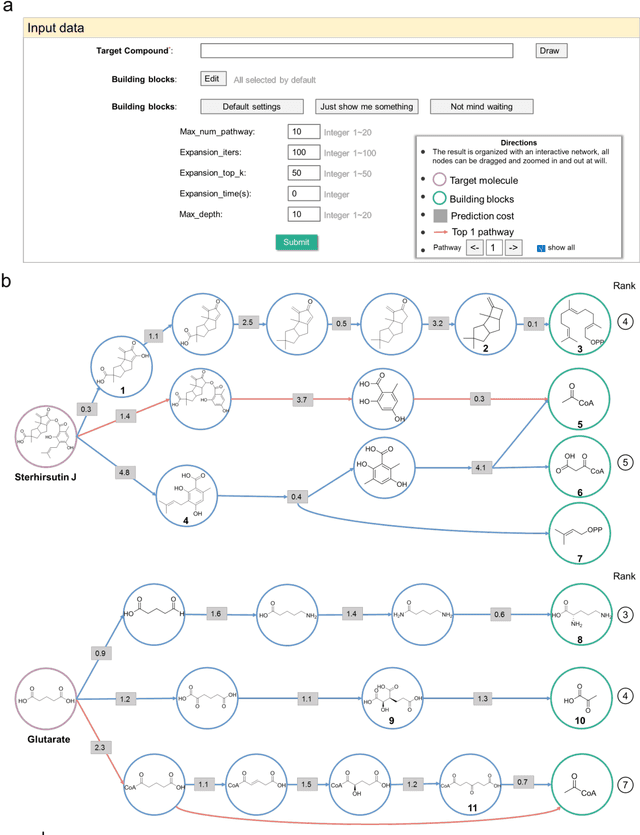
Abstract:Nature, a synthetic master, creates more than 300,000 natural products (NPs) which are the major constituents of FDA-proved drugs owing to the vast chemical space of NPs. To date, there are fewer than 30,000 validated NPs compounds involved in about 33,000 known enzyme catalytic reactions, and even fewer biosynthetic pathways are known with complete cascade-connected enzyme catalysis. Therefore, it is valuable to make computer-aided bio-retrosynthesis predictions. Here, we develop BioNavi-NP, a navigable and user-friendly toolkit, which is capable of predicting the biosynthetic pathways for NPs and NP-like compounds through a novel (AND-OR Tree)-based planning algorithm, an enhanced molecular Transformer neural network, and a training set that combines general organic transformations and biosynthetic steps. Extensive evaluations reveal that BioNavi-NP generalizes well to identifying the reported biosynthetic pathways for 90% of test compounds and recovering the verified building blocks for 73%, significantly outperforming conventional rule-based approaches. Moreover, BioNavi-NP also shows an outstanding capacity of biologically plausible pathways enumeration. In this sense, BioNavi-NP is a leading-edge toolkit to redesign complex biosynthetic pathways of natural products with applications to total or semi-synthesis and pathway elucidation or reconstruction.
Speeding up Computational Morphogenesis with Online Neural Synthetic Gradients
Apr 27, 2021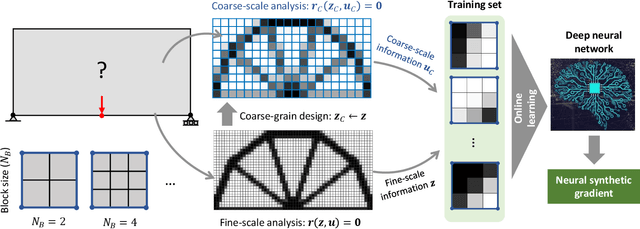
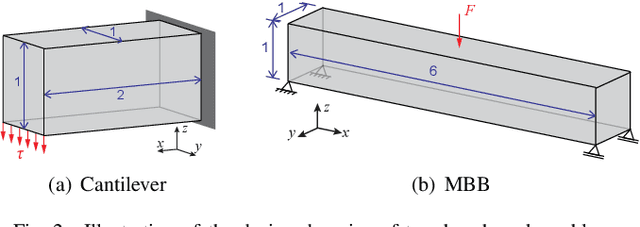
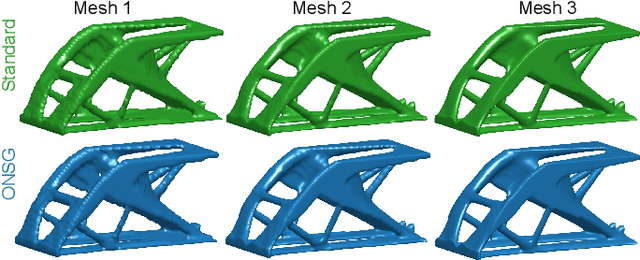
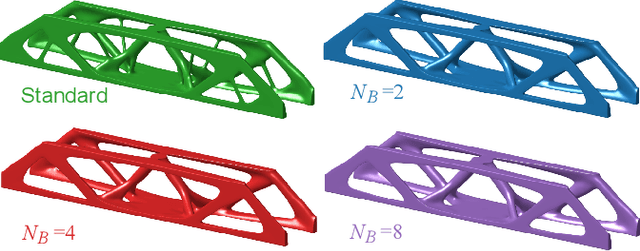
Abstract:A wide range of modern science and engineering applications are formulated as optimization problems with a system of partial differential equations (PDEs) as constraints. These PDE-constrained optimization problems are typically solved in a standard discretize-then-optimize approach. In many industry applications that require high-resolution solutions, the discretized constraints can easily have millions or even billions of variables, making it very slow for the standard iterative optimizer to solve the exact gradients. In this work, we propose a general framework to speed up PDE-constrained optimization using online neural synthetic gradients (ONSG) with a novel two-scale optimization scheme. We successfully apply our ONSG framework to computational morphogenesis, a representative and challenging class of PDE-constrained optimization problems. Extensive experiments have demonstrated that our method can significantly speed up computational morphogenesis (also known as topology optimization), and meanwhile maintain the quality of final solution compared to the standard optimizer. On a large-scale 3D optimal design problem with around 1,400,000 design variables, our method achieves up to 7.5x speedup while producing optimized designs with comparable objectives.
How to Design Sample and Computationally Efficient VQA Models
Mar 22, 2021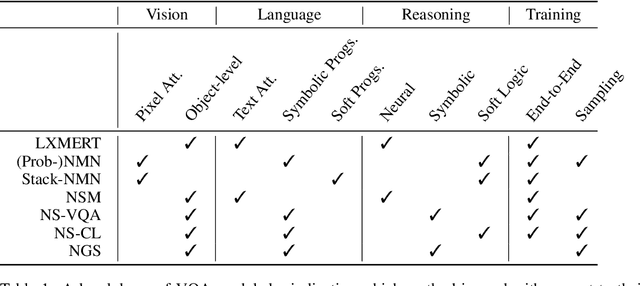
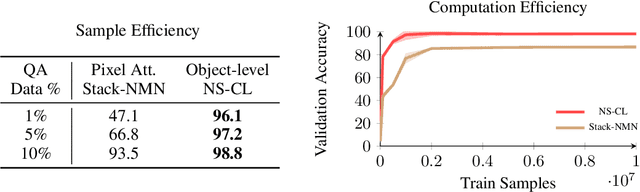
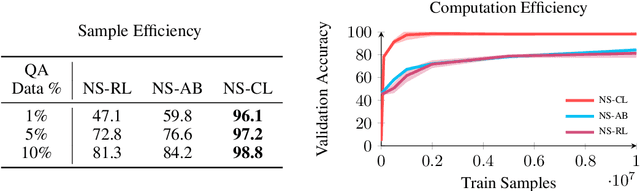
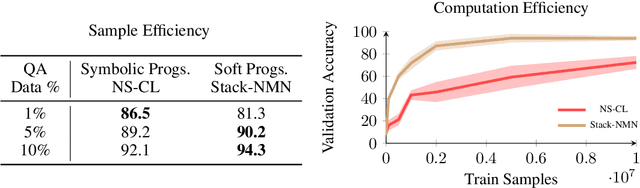
Abstract:In multi-modal reasoning tasks, such as visual question answering (VQA), there have been many modeling and training paradigms tested. Previous models propose different methods for the vision and language tasks, but which ones perform the best while being sample and computationally efficient? Based on our experiments, we find that representing the text as probabilistic programs and images as object-level scene graphs best satisfy these desiderata. We extend existing models to leverage these soft programs and scene graphs to train on question answer pairs in an end-to-end manner. Empirical results demonstrate that this differentiable end-to-end program executor is able to maintain state-of-the-art accuracy while being sample and computationally efficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge