Image To Image Translation
Image-to-image translation is the process of converting an image from one domain to another using deep learning techniques.
Papers and Code
Theoretical Bounds on Parallel Imaging Implicit Data Crimes in an MRI Reproducing Kernel Hilbert Space
Nov 19, 2025Magnetic Resonance Imaging (MRI) diagnoses and manages a wide range of diseases, yet long scan times drive high costs and limit accessibility. AI methods have demonstrated substantial potential for reducing scan times, but despite rapid progress, clinical translation of AI often fails. One particular class of failure modes, referred to as implicit data crimes, are a result of hidden biases introduced when MRI datasets incompletely model the MRI physics of the acquisition. Previous work identified data crimes resulting from algorithmic completion of k-space with parallel imaging and drew on simulation to demonstrate the resulting downstream biases. This work proposes a mathematical framework to re-characterize the problem as one of error reduction during interpolation between sets of evaluation coordinates. We establish a generalized matrix-based definition of the reconstruction error upper bound as a function of the input sampling pattern. Experiments on relevant sampling pattern structures demonstrate the relevance of the framework and suggest future directions for analysis of data crimes.
US-X Complete: A Multi-Modal Approach to Anatomical 3D Shape Recovery
Nov 19, 2025Ultrasound offers a radiation-free, cost-effective solution for real-time visualization of spinal landmarks, paraspinal soft tissues and neurovascular structures, making it valuable for intraoperative guidance during spinal procedures. However, ultrasound suffers from inherent limitations in visualizing complete vertebral anatomy, in particular vertebral bodies, due to acoustic shadowing effects caused by bone. In this work, we present a novel multi-modal deep learning method for completing occluded anatomical structures in 3D ultrasound by leveraging complementary information from a single X-ray image. To enable training, we generate paired training data consisting of: (1) 2D lateral vertebral views that simulate X-ray scans, and (2) 3D partial vertebrae representations that mimic the limited visibility and occlusions encountered during ultrasound spine imaging. Our method integrates morphological information from both imaging modalities and demonstrates significant improvements in vertebral reconstruction (p < 0.001) compared to state of art in 3D ultrasound vertebral completion. We perform phantom studies as an initial step to future clinical translation, and achieve a more accurate, complete volumetric lumbar spine visualization overlayed on the ultrasound scan without the need for registration with preoperative modalities such as computed tomography. This demonstrates that integrating a single X-ray projection mitigates ultrasound's key limitation while preserving its strengths as the primary imaging modality. Code and data can be found at https://github.com/miruna20/US-X-Complete
A Specialized Large Language Model for Clinical Reasoning and Diagnosis in Rare Diseases
Nov 18, 2025Rare diseases affect hundreds of millions worldwide, yet diagnosis often spans years. Convectional pipelines decouple noisy evidence extraction from downstream inferential diagnosis, and general/medical large language models (LLMs) face scarce real world electronic health records (EHRs), stale domain knowledge, and hallucinations. We assemble a large, domain specialized clinical corpus and a clinician validated reasoning set, and develop RareSeek R1 via staged instruction tuning, chain of thought learning, and graph grounded retrieval. Across multicenter EHR narratives and public benchmarks, RareSeek R1 attains state of the art accuracy, robust generalization, and stability under noisy or overlapping phenotypes. Augmented retrieval yields the largest gains when narratives pair with prioritized variants by resolving ambiguity and aligning candidates to mechanisms. Human studies show performance on par with experienced physicians and consistent gains in assistive use. Notably, transparent reasoning highlights decisive non phenotypic evidence (median 23.1%, such as imaging, interventions, functional tests) underpinning many correct diagnoses. This work advances a narrative first, knowledge integrated reasoning paradigm that shortens the diagnostic odyssey and enables auditable, clinically translatable decision support.
Free-Form Scene Editor: Enabling Multi-Round Object Manipulation like in a 3D Engine
Nov 17, 2025Recent advances in text-to-image (T2I) diffusion models have significantly improved semantic image editing, yet most methods fall short in performing 3D-aware object manipulation. In this work, we present FFSE, a 3D-aware autoregressive framework designed to enable intuitive, physically-consistent object editing directly on real-world images. Unlike previous approaches that either operate in image space or require slow and error-prone 3D reconstruction, FFSE models editing as a sequence of learned 3D transformations, allowing users to perform arbitrary manipulations, such as translation, scaling, and rotation, while preserving realistic background effects (e.g., shadows, reflections) and maintaining global scene consistency across multiple editing rounds. To support learning of multi-round 3D-aware object manipulation, we introduce 3DObjectEditor, a hybrid dataset constructed from simulated editing sequences across diverse objects and scenes, enabling effective training under multi-round and dynamic conditions. Extensive experiments show that the proposed FFSE significantly outperforms existing methods in both single-round and multi-round 3D-aware editing scenarios.
MRIQT: Physics-Aware Diffusion Model for Image Quality Transfer in Neonatal Ultra-Low-Field MRI
Nov 17, 2025Portable ultra-low-field MRI (uLF-MRI, 0.064 T) offers accessible neuroimaging for neonatal care but suffers from low signal-to-noise ratio and poor diagnostic quality compared to high-field (HF) MRI. We propose MRIQT, a 3D conditional diffusion framework for image quality transfer (IQT) from uLF to HF MRI. MRIQT combines realistic K-space degradation for physics-consistent uLF simulation, v-prediction with classifier-free guidance for stable image-to-image generation, and an SNR-weighted 3D perceptual loss for anatomical fidelity. The model denoises from a noised uLF input conditioned on the same scan, leveraging volumetric attention-UNet architecture for structure-preserving translation. Trained on a neonatal cohort with diverse pathologies, MRIQT surpasses recent GAN and CNN baselines in PSNR 15.3% with 1.78% over the state of the art, while physicians rated 85% of its outputs as good quality with clear pathology present. MRIQT enables high-fidelity, diffusion-based enhancement of portable ultra-low-field (uLF) MRI for deliable neonatal brain assessment.
From Retinal Pixels to Patients: Evolution of Deep Learning Research in Diabetic Retinopathy Screening
Nov 14, 2025



Diabetic Retinopathy (DR) remains a leading cause of preventable blindness, with early detection critical for reducing vision loss worldwide. Over the past decade, deep learning has transformed DR screening, progressing from early convolutional neural networks trained on private datasets to advanced pipelines addressing class imbalance, label scarcity, domain shift, and interpretability. This survey provides the first systematic synthesis of DR research spanning 2016-2025, consolidating results from 50+ studies and over 20 datasets. We critically examine methodological advances, including self- and semi-supervised learning, domain generalization, federated training, and hybrid neuro-symbolic models, alongside evaluation protocols, reporting standards, and reproducibility challenges. Benchmark tables contextualize performance across datasets, while discussion highlights open gaps in multi-center validation and clinical trust. By linking technical progress with translational barriers, this work outlines a practical agenda for reproducible, privacy-preserving, and clinically deployable DR AI. Beyond DR, many of the surveyed innovations extend broadly to medical imaging at scale.
GRLoc: Geometric Representation Regression for Visual Localization
Nov 17, 2025



Absolute Pose Regression (APR) has emerged as a compelling paradigm for visual localization. However, APR models typically operate as black boxes, directly regressing a 6-DoF pose from a query image, which can lead to memorizing training views rather than understanding 3D scene geometry. In this work, we propose a geometrically-grounded alternative. Inspired by novel view synthesis, which renders images from intermediate geometric representations, we reformulate APR as its inverse that regresses the underlying 3D representations directly from the image, and we name this paradigm Geometric Representation Regression (GRR). Our model explicitly predicts two disentangled geometric representations in the world coordinate system: (1) a ray bundle's directions to estimate camera rotation, and (2) a corresponding pointmap to estimate camera translation. The final 6-DoF camera pose is then recovered from these geometric components using a differentiable deterministic solver. This disentangled approach, which separates the learned visual-to-geometry mapping from the final pose calculation, introduces a strong geometric prior into the network. We find that the explicit decoupling of rotation and translation predictions measurably boosts performance. We demonstrate state-of-the-art performance on 7-Scenes and Cambridge Landmarks datasets, validating that modeling the inverse rendering process is a more robust path toward generalizable absolute pose estimation.
LINGUAL: Language-INtegrated GUidance in Active Learning for Medical Image Segmentation
Nov 18, 2025Although active learning (AL) in segmentation tasks enables experts to annotate selected regions of interest (ROIs) instead of entire images, it remains highly challenging, labor-intensive, and cognitively demanding due to the blurry and ambiguous boundaries commonly observed in medical images. Also, in conventional AL, annotation effort is a function of the ROI- larger regions make the task cognitively easier but incur higher annotation costs, whereas smaller regions demand finer precision and more attention from the expert. In this context, language guidance provides an effective alternative, requiring minimal expert effort while bypassing the cognitively demanding task of precise boundary delineation in segmentation. Towards this goal, we introduce LINGUAL: a framework that receives natural language instructions from an expert, translates them into executable programs through in-context learning, and automatically performs the corresponding sequence of sub-tasks without any human intervention. We demonstrate the effectiveness of LINGUAL in active domain adaptation (ADA) achieving comparable or superior performance to AL baselines while reducing estimated annotation time by approximately 80%.
Towards High-Consistency Embodied World Model with Multi-View Trajectory Videos
Nov 19, 2025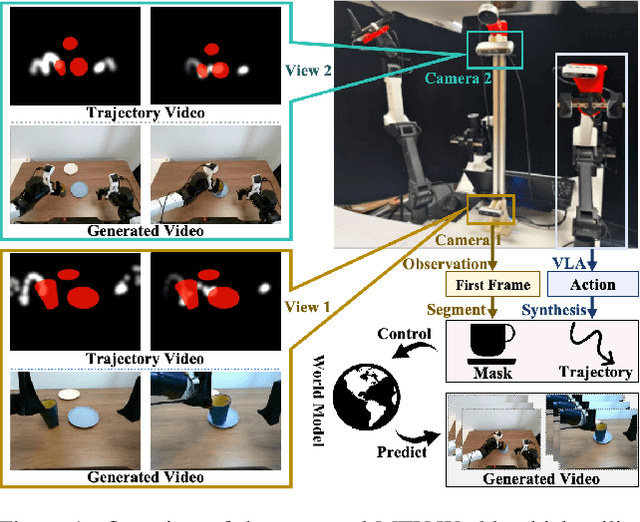
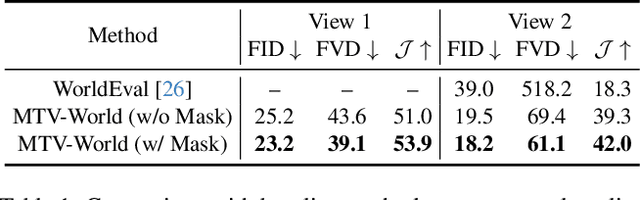
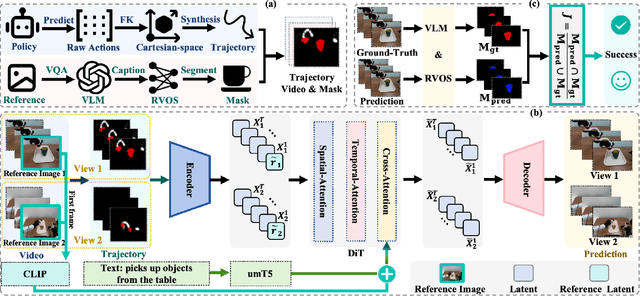
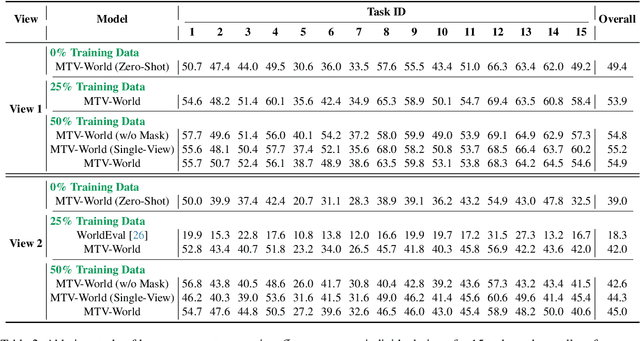
Embodied world models aim to predict and interact with the physical world through visual observations and actions. However, existing models struggle to accurately translate low-level actions (e.g., joint positions) into precise robotic movements in predicted frames, leading to inconsistencies with real-world physical interactions. To address these limitations, we propose MTV-World, an embodied world model that introduces Multi-view Trajectory-Video control for precise visuomotor prediction. Specifically, instead of directly using low-level actions for control, we employ trajectory videos obtained through camera intrinsic and extrinsic parameters and Cartesian-space transformation as control signals. However, projecting 3D raw actions onto 2D images inevitably causes a loss of spatial information, making a single view insufficient for accurate interaction modeling. To overcome this, we introduce a multi-view framework that compensates for spatial information loss and ensures high-consistency with physical world. MTV-World forecasts future frames based on multi-view trajectory videos as input and conditioning on an initial frame per view. Furthermore, to systematically evaluate both robotic motion precision and object interaction accuracy, we develop an auto-evaluation pipeline leveraging multimodal large models and referring video object segmentation models. To measure spatial consistency, we formulate it as an object location matching problem and adopt the Jaccard Index as the evaluation metric. Extensive experiments demonstrate that MTV-World achieves precise control execution and accurate physical interaction modeling in complex dual-arm scenarios.
nnMIL: A generalizable multiple instance learning framework for computational pathology
Nov 18, 2025Computational pathology holds substantial promise for improving diagnosis and guiding treatment decisions. Recent pathology foundation models enable the extraction of rich patch-level representations from large-scale whole-slide images (WSIs), but current approaches for aggregating these features into slide-level predictions remain constrained by design limitations that hinder generalizability and reliability. Here, we developed nnMIL, a simple yet broadly applicable multiple-instance learning framework that connects patch-level foundation models to robust slide-level clinical inference. nnMIL introduces random sampling at both the patch and feature levels, enabling large-batch optimization, task-aware sampling strategies, and efficient and scalable training across datasets and model architectures. A lightweight aggregator performs sliding-window inference to generate ensemble slide-level predictions and supports principled uncertainty estimation. Across 40,000 WSIs encompassing 35 clinical tasks and four pathology foundation models, nnMIL consistently outperformed existing MIL methods for disease diagnosis, histologic subtyping, molecular biomarker detection, and pan- cancer prognosis prediction. It further demonstrated strong cross-model generalization, reliable uncertainty quantification, and robust survival stratification in multiple external cohorts. In conclusion, nnMIL offers a practical and generalizable solution for translating pathology foundation models into clinically meaningful predictions, advancing the development and deployment of reliable AI systems in real-world settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge