Image Registration
Image registration is the process of transforming different sets of data into one coordinate system. Data may be multiple photographs, data from different sensors, times, depths, or viewpoints. It is used in computer vision, medical imaging, and compiling and analyzing images and data from satellites. Registration is necessary in order to be able to compare or integrate the data obtained from these different measurements.
Papers and Code
PADReg: Physics-Aware Deformable Registration Guided by Contact Force for Ultrasound Sequences
Aug 12, 2025Ultrasound deformable registration estimates spatial transformations between pairs of deformed ultrasound images, which is crucial for capturing biomechanical properties and enhancing diagnostic accuracy in diseases such as thyroid nodules and breast cancer. However, ultrasound deformable registration remains highly challenging, especially under large deformation. The inherently low contrast, heavy noise and ambiguous tissue boundaries in ultrasound images severely hinder reliable feature extraction and correspondence matching. Existing methods often suffer from poor anatomical alignment and lack physical interpretability. To address the problem, we propose PADReg, a physics-aware deformable registration framework guided by contact force. PADReg leverages synchronized contact force measured by robotic ultrasound systems as a physical prior to constrain the registration. Specifically, instead of directly predicting deformation fields, we first construct a pixel-wise stiffness map utilizing the multi-modal information from contact force and ultrasound images. The stiffness map is then combined with force data to estimate a dense deformation field, through a lightweight physics-aware module inspired by Hooke's law. This design enables PADReg to achieve physically plausible registration with better anatomical alignment than previous methods relying solely on image similarity. Experiments on in-vivo datasets demonstrate that it attains a HD95 of 12.90, which is 21.34\% better than state-of-the-art methods. The source code is available at https://github.com/evelynskip/PADReg.
Modality-Aware Feature Matching: A Comprehensive Review of Single- and Cross-Modality Techniques
Jul 30, 2025



Feature matching is a cornerstone task in computer vision, essential for applications such as image retrieval, stereo matching, 3D reconstruction, and SLAM. This survey comprehensively reviews modality-based feature matching, exploring traditional handcrafted methods and emphasizing contemporary deep learning approaches across various modalities, including RGB images, depth images, 3D point clouds, LiDAR scans, medical images, and vision-language interactions. Traditional methods, leveraging detectors like Harris corners and descriptors such as SIFT and ORB, demonstrate robustness under moderate intra-modality variations but struggle with significant modality gaps. Contemporary deep learning-based methods, exemplified by detector-free strategies like CNN-based SuperPoint and transformer-based LoFTR, substantially improve robustness and adaptability across modalities. We highlight modality-aware advancements, such as geometric and depth-specific descriptors for depth images, sparse and dense learning methods for 3D point clouds, attention-enhanced neural networks for LiDAR scans, and specialized solutions like the MIND descriptor for complex medical image matching. Cross-modal applications, particularly in medical image registration and vision-language tasks, underscore the evolution of feature matching to handle increasingly diverse data interactions.
Large-scale Multi-sequence Pretraining for Generalizable MRI Analysis in Versatile Clinical Applications
Aug 10, 2025



Multi-sequence Magnetic Resonance Imaging (MRI) offers remarkable versatility, enabling the distinct visualization of different tissue types. Nevertheless, the inherent heterogeneity among MRI sequences poses significant challenges to the generalization capability of deep learning models. These challenges undermine model performance when faced with varying acquisition parameters, thereby severely restricting their clinical utility. In this study, we present PRISM, a foundation model PRe-trained with large-scale multI-Sequence MRI. We collected a total of 64 datasets from both public and private sources, encompassing a wide range of whole-body anatomical structures, with scans spanning diverse MRI sequences. Among them, 336,476 volumetric MRI scans from 34 datasets (8 public and 26 private) were curated to construct the largest multi-organ multi-sequence MRI pretraining corpus to date. We propose a novel pretraining paradigm that disentangles anatomically invariant features from sequence-specific variations in MRI, while preserving high-level semantic representations. We established a benchmark comprising 44 downstream tasks, including disease diagnosis, image segmentation, registration, progression prediction, and report generation. These tasks were evaluated on 32 public datasets and 5 private cohorts. PRISM consistently outperformed both non-pretrained models and existing foundation models, achieving first-rank results in 39 out of 44 downstream benchmarks with statistical significance improvements. These results underscore its ability to learn robust and generalizable representations across unseen data acquired under diverse MRI protocols. PRISM provides a scalable framework for multi-sequence MRI analysis, thereby enhancing the translational potential of AI in radiology. It delivers consistent performance across diverse imaging protocols, reinforcing its clinical applicability.
DiffVL: Diffusion-Based Visual Localization on 2D Maps via BEV-Conditioned GPS Denoising
Sep 18, 2025



Accurate visual localization is crucial for autonomous driving, yet existing methods face a fundamental dilemma: While high-definition (HD) maps provide high-precision localization references, their costly construction and maintenance hinder scalability, which drives research toward standard-definition (SD) maps like OpenStreetMap. Current SD-map-based approaches primarily focus on Bird's-Eye View (BEV) matching between images and maps, overlooking a ubiquitous signal-noisy GPS. Although GPS is readily available, it suffers from multipath errors in urban environments. We propose DiffVL, the first framework to reformulate visual localization as a GPS denoising task using diffusion models. Our key insight is that noisy GPS trajectory, when conditioned on visual BEV features and SD maps, implicitly encode the true pose distribution, which can be recovered through iterative diffusion refinement. DiffVL, unlike prior BEV-matching methods (e.g., OrienterNet) or transformer-based registration approaches, learns to reverse GPS noise perturbations by jointly modeling GPS, SD map, and visual signals, achieving sub-meter accuracy without relying on HD maps. Experiments on multiple datasets demonstrate that our method achieves state-of-the-art accuracy compared to BEV-matching baselines. Crucially, our work proves that diffusion models can enable scalable localization by treating noisy GPS as a generative prior-making a paradigm shift from traditional matching-based methods.
A Registration-Based Star-Shape Segmentation Model and Fast Algorithms
Aug 11, 2025Image segmentation plays a crucial role in extracting objects of interest and identifying their boundaries within an image. However, accurate segmentation becomes challenging when dealing with occlusions, obscurities, or noise in corrupted images. To tackle this challenge, prior information is often utilized, with recent attention on star-shape priors. In this paper, we propose a star-shape segmentation model based on the registration framework. By combining the level set representation with the registration framework and imposing constraints on the deformed level set function, our model enables both full and partial star-shape segmentation, accommodating single or multiple centers. Additionally, our approach allows for the enforcement of identified boundaries to pass through specified landmark locations. We tackle the proposed models using the alternating direction method of multipliers. Through numerical experiments conducted on synthetic and real images, we demonstrate the efficacy of our approach in achieving accurate star-shape segmentation.
fastWDM3D: Fast and Accurate 3D Healthy Tissue Inpainting
Jul 17, 2025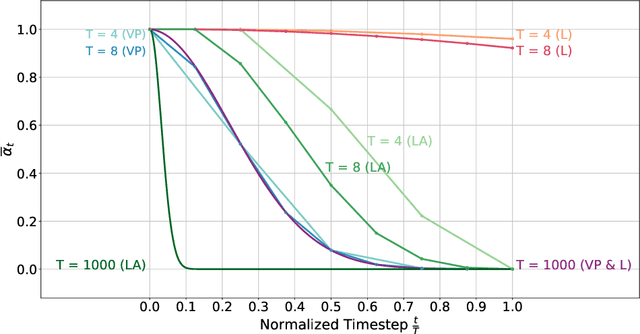
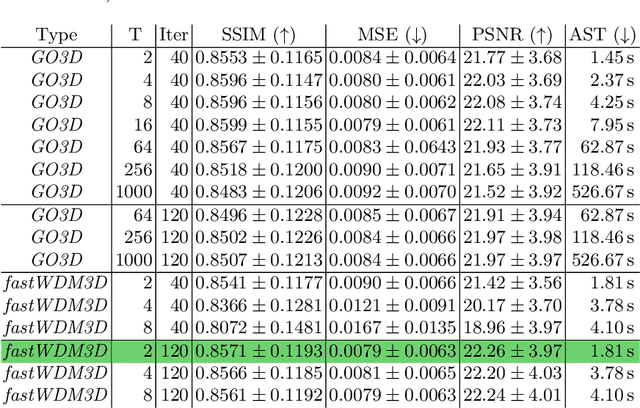
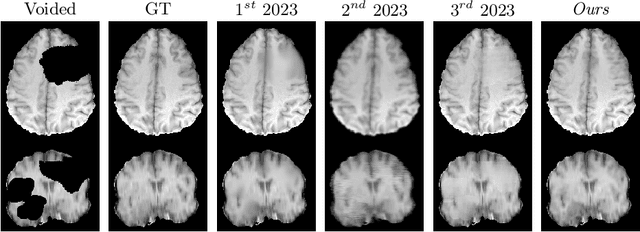
Healthy tissue inpainting has significant applications, including the generation of pseudo-healthy baselines for tumor growth models and the facilitation of image registration. In previous editions of the BraTS Local Synthesis of Healthy Brain Tissue via Inpainting Challenge, denoising diffusion probabilistic models (DDPMs) demonstrated qualitatively convincing results but suffered from low sampling speed. To mitigate this limitation, we adapted a 2D image generation approach, combining DDPMs with generative adversarial networks (GANs) and employing a variance-preserving noise schedule, for the task of 3D inpainting. Our experiments showed that the variance-preserving noise schedule and the selected reconstruction losses can be effectively utilized for high-quality 3D inpainting in a few time steps without requiring adversarial training. We applied our findings to a different architecture, a 3D wavelet diffusion model (WDM3D) that does not include a GAN component. The resulting model, denoted as fastWDM3D, obtained a SSIM of 0.8571, a MSE of 0.0079, and a PSNR of 22.26 on the BraTS inpainting test set. Remarkably, it achieved these scores using only two time steps, completing the 3D inpainting process in 1.81 s per image. When compared to other DDPMs used for healthy brain tissue inpainting, our model is up to 800 x faster while still achieving superior performance metrics. Our proposed method, fastWDM3D, represents a promising approach for fast and accurate healthy tissue inpainting. Our code is available at https://github.com/AliciaDurrer/fastWDM3D.
Deformable Medical Image Registration with Effective Anatomical Structure Representation and Divide-and-Conquer Network
Jun 24, 2025Effective representation of Regions of Interest (ROI) and independent alignment of these ROIs can significantly enhance the performance of deformable medical image registration (DMIR). However, current learning-based DMIR methods have limitations. Unsupervised techniques disregard ROI representation and proceed directly with aligning pairs of images, while weakly-supervised methods heavily depend on label constraints to facilitate registration. To address these issues, we introduce a novel ROI-based registration approach named EASR-DCN. Our method represents medical images through effective ROIs and achieves independent alignment of these ROIs without requiring labels. Specifically, we first used a Gaussian mixture model for intensity analysis to represent images using multiple effective ROIs with distinct intensities. Furthermore, we propose a novel Divide-and-Conquer Network (DCN) to process these ROIs through separate channels to learn feature alignments for each ROI. The resultant correspondences are seamlessly integrated to generate a comprehensive displacement vector field. Extensive experiments were performed on three MRI and one CT datasets to showcase the superior accuracy and deformation reduction efficacy of our EASR-DCN. Compared to VoxelMorph, our EASR-DCN achieved improvements of 10.31\% in the Dice score for brain MRI, 13.01\% for cardiac MRI, and 5.75\% for hippocampus MRI, highlighting its promising potential for clinical applications. The code for this work will be released upon acceptance of the paper.
Shape Completion and Real-Time Visualization in Robotic Ultrasound Spine Acquisitions
Aug 12, 2025



Ultrasound (US) imaging is increasingly used in spinal procedures due to its real-time, radiation-free capabilities; however, its effectiveness is hindered by shadowing artifacts that obscure deeper tissue structures. Traditional approaches, such as CT-to-US registration, incorporate anatomical information from preoperative CT scans to guide interventions, but they are limited by complex registration requirements, differences in spine curvature, and the need for recent CT imaging. Recent shape completion methods can offer an alternative by reconstructing spinal structures in US data, while being pretrained on large set of publicly available CT scans. However, these approaches are typically offline and have limited reproducibility. In this work, we introduce a novel integrated system that combines robotic ultrasound with real-time shape completion to enhance spinal visualization. Our robotic platform autonomously acquires US sweeps of the lumbar spine, extracts vertebral surfaces from ultrasound, and reconstructs the complete anatomy using a deep learning-based shape completion network. This framework provides interactive, real-time visualization with the capability to autonomously repeat scans and can enable navigation to target locations. This can contribute to better consistency, reproducibility, and understanding of the underlying anatomy. We validate our approach through quantitative experiments assessing shape completion accuracy and evaluations of multiple spine acquisition protocols on a phantom setup. Additionally, we present qualitative results of the visualization on a volunteer scan.
Explainable AI for Collaborative Assessment of 2D/3D Registration Quality
Jul 23, 2025As surgery embraces digital transformation--integrating sophisticated imaging, advanced algorithms, and robotics to support and automate complex sub-tasks--human judgment of system correctness remains a vital safeguard for patient safety. This shift introduces new "operator-type" roles tasked with verifying complex algorithmic outputs, particularly at critical junctures of the procedure, such as the intermediary check before drilling or implant placement. A prime example is 2D/3D registration, a key enabler of image-based surgical navigation that aligns intraoperative 2D images with preoperative 3D data. Although registration algorithms have advanced significantly, they occasionally yield inaccurate results. Because even small misalignments can lead to revision surgery or irreversible surgical errors, there is a critical need for robust quality assurance. Current visualization-based strategies alone have been found insufficient to enable humans to reliably detect 2D/3D registration misalignments. In response, we propose the first artificial intelligence (AI) framework trained specifically for 2D/3D registration quality verification, augmented by explainability features that clarify the model's decision-making. Our explainable AI (XAI) approach aims to enhance informed decision-making for human operators by providing a second opinion together with a rationale behind it. Through algorithm-centric and human-centered evaluations, we systematically compare four conditions: AI-only, human-only, human-AI, and human-XAI. Our findings reveal that while explainability features modestly improve user trust and willingness to override AI errors, they do not exceed the standalone AI in aggregate performance. Nevertheless, future work extending both the algorithmic design and the human-XAI collaboration elements holds promise for more robust quality assurance of 2D/3D registration.
Diff$^2$I2P: Differentiable Image-to-Point Cloud Registration with Diffusion Prior
Jul 09, 2025Learning cross-modal correspondences is essential for image-to-point cloud (I2P) registration. Existing methods achieve this mostly by utilizing metric learning to enforce feature alignment across modalities, disregarding the inherent modality gap between image and point data. Consequently, this paradigm struggles to ensure accurate cross-modal correspondences. To this end, inspired by the cross-modal generation success of recent large diffusion models, we propose Diff$^2$I2P, a fully Differentiable I2P registration framework, leveraging a novel and effective Diffusion prior for bridging the modality gap. Specifically, we propose a Control-Side Score Distillation (CSD) technique to distill knowledge from a depth-conditioned diffusion model to directly optimize the predicted transformation. However, the gradients on the transformation fail to backpropagate onto the cross-modal features due to the non-differentiability of correspondence retrieval and PnP solver. To this end, we further propose a Deformable Correspondence Tuning (DCT) module to estimate the correspondences in a differentiable way, followed by the transformation estimation using a differentiable PnP solver. With these two designs, the Diffusion model serves as a strong prior to guide the cross-modal feature learning of image and point cloud for forming robust correspondences, which significantly improves the registration. Extensive experimental results demonstrate that Diff$^2$I2P consistently outperforms SoTA I2P registration methods, achieving over 7% improvement in registration recall on the 7-Scenes benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge