Yihang Wu
Federated Vision Transformer with Adaptive Focal Loss for Medical Image Classification
Feb 02, 2026Abstract:While deep learning models like Vision Transformer (ViT) have achieved significant advances, they typically require large datasets. With data privacy regulations, access to many original datasets is restricted, especially medical images. Federated learning (FL) addresses this challenge by enabling global model aggregation without data exchange. However, the heterogeneity of the data and the class imbalance that exist in local clients pose challenges for the generalization of the model. This study proposes a FL framework leveraging a dynamic adaptive focal loss (DAFL) and a client-aware aggregation strategy for local training. Specifically, we design a dynamic class imbalance coefficient that adjusts based on each client's sample distribution and class data distribution, ensuring minority classes receive sufficient attention and preventing sparse data from being ignored. To address client heterogeneity, a weighted aggregation strategy is adopted, which adapts to data size and characteristics to better capture inter-client variations. The classification results on three public datasets (ISIC, Ocular Disease and RSNA-ICH) show that the proposed framework outperforms DenseNet121, ResNet50, ViT-S/16, ViT-L/32, FedCLIP, Swin Transformer, CoAtNet, and MixNet in most cases, with accuracy improvements ranging from 0.98\% to 41.69\%. Ablation studies on the imbalanced ISIC dataset validate the effectiveness of the proposed loss function and aggregation strategy compared to traditional loss functions and other FL approaches. The codes can be found at: https://github.com/AIPMLab/ViT-FLDAF.
Do AI Overviews Benefit Search Engines? An Ecosystem Perspective
Jan 30, 2026Abstract:The integration of AI Overviews into search engines enhances user experience but diverts traffic from content creators, potentially discouraging high-quality content creation and causing user attrition that undermines long-term search engine profit. To address this issue, we propose a game-theoretic model of creator competition with costly effort, characterize equilibrium behavior, and design two incentive mechanisms: a citation mechanism that references sources within an AI Overview, and a compensation mechanism that offers monetary rewards to creators. For both cases, we provide structural insights and near-optimal profit-maximizing mechanisms. Evaluations on real click data show that although AI Overviews harm long-term search engine profit, interventions based on our proposed mechanisms can increase long-term profit across a range of realistic scenarios, pointing toward a more sustainable trajectory for AI-enhanced search ecosystems.
Reliable and Private Utility Signaling for Data Markets
Nov 11, 2025Abstract:The explosive growth of data has highlighted its critical role in driving economic growth through data marketplaces, which enable extensive data sharing and access to high-quality datasets. To support effective trading, signaling mechanisms provide participants with information about data products before transactions, enabling informed decisions and facilitating trading. However, due to the inherent free-duplication nature of data, commonly practiced signaling methods face a dilemma between privacy and reliability, undermining the effectiveness of signals in guiding decision-making. To address this, this paper explores the benefits and develops a non-TCP-based construction for a desirable signaling mechanism that simultaneously ensures privacy and reliability. We begin by formally defining the desirable utility signaling mechanism and proving its ability to prevent suboptimal decisions for both participants and facilitate informed data trading. To design a protocol to realize its functionality, we propose leveraging maliciously secure multi-party computation (MPC) to ensure the privacy and robustness of signal computation and introduce an MPC-based hash verification scheme to ensure input reliability. In multi-seller scenarios requiring fair data valuation, we further explore the design and optimization of the MPC-based KNN-Shapley method with improved efficiency. Rigorous experiments demonstrate the efficiency and practicality of our approach.
Federated CLIP for Resource-Efficient Heterogeneous Medical Image Classification
Nov 11, 2025Abstract:Despite the remarkable performance of deep models in medical imaging, they still require source data for training, which limits their potential in light of privacy concerns. Federated learning (FL), as a decentralized learning framework that trains a shared model with multiple hospitals (a.k.a., FL clients), provides a feasible solution. However, data heterogeneity and resource costs hinder the deployment of FL models, especially when using vision language models (VLM). To address these challenges, we propose a novel contrastive language-image pre-training (CLIP) based FL approach for medical image classification (FedMedCLIP). Specifically, we introduce a masked feature adaptation module (FAM) as a communication module to reduce the communication load while freezing the CLIP encoders to reduce the computational overhead. Furthermore, we propose a masked multi-layer perceptron (MLP) as a private local classifier to adapt to the client tasks. Moreover, we design an adaptive Kullback-Leibler (KL) divergence-based distillation regularization method to enable mutual learning between FAM and MLP. Finally, we incorporate model compression to transmit the FAM parameters while using ensemble predictions for classification. Extensive experiments on four publicly available medical datasets demonstrate that our model provides feasible performance (e.g., 8\% higher compared to second best baseline on ISIC2019) with reasonable resource cost (e.g., 120$\times$ faster than FedAVG).
Breaking the Modality Barrier: Generative Modeling for Accurate Molecule Retrieval from Mass Spectra
Nov 09, 2025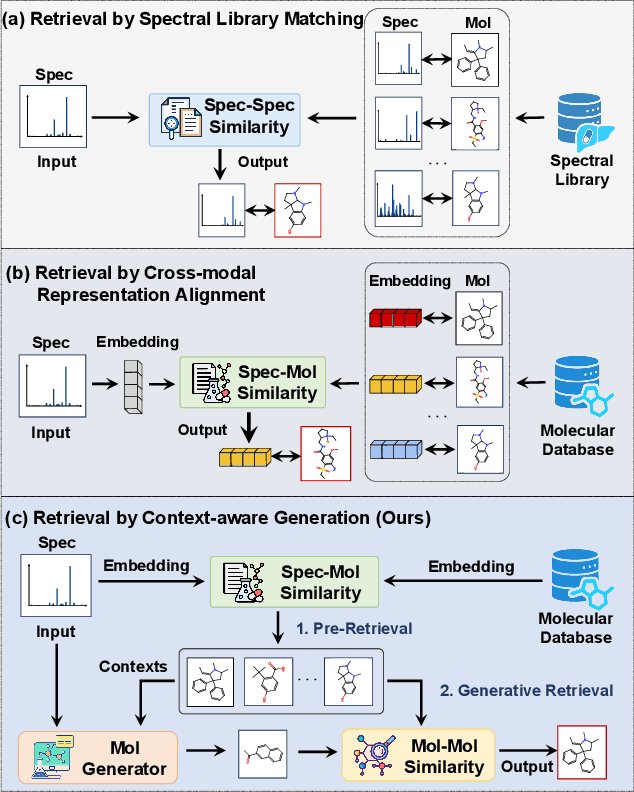



Abstract:Retrieving molecular structures from tandem mass spectra is a crucial step in rapid compound identification. Existing retrieval methods, such as traditional mass spectral library matching, suffer from limited spectral library coverage, while recent cross-modal representation learning frameworks often encounter modality misalignment, resulting in suboptimal retrieval accuracy and generalization. To address these limitations, we propose GLMR, a Generative Language Model-based Retrieval framework that mitigates the cross-modal misalignment through a two-stage process. In the pre-retrieval stage, a contrastive learning-based model identifies top candidate molecules as contextual priors for the input mass spectrum. In the generative retrieval stage, these candidate molecules are integrated with the input mass spectrum to guide a generative model in producing refined molecular structures, which are then used to re-rank the candidates based on molecular similarity. Experiments on both MassSpecGym and the proposed MassRET-20k dataset demonstrate that GLMR significantly outperforms existing methods, achieving over 40% improvement in top-1 accuracy and exhibiting strong generalizability.
Enhancing Dual Network Based Semi-Supervised Medical Image Segmentation with Uncertainty-Guided Pseudo-Labeling
Sep 16, 2025Abstract:Despite the remarkable performance of supervised medical image segmentation models, relying on a large amount of labeled data is impractical in real-world situations. Semi-supervised learning approaches aim to alleviate this challenge using unlabeled data through pseudo-label generation. Yet, existing semi-supervised segmentation methods still suffer from noisy pseudo-labels and insufficient supervision within the feature space. To solve these challenges, this paper proposes a novel semi-supervised 3D medical image segmentation framework based on a dual-network architecture. Specifically, we investigate a Cross Consistency Enhancement module using both cross pseudo and entropy-filtered supervision to reduce the noisy pseudo-labels, while we design a dynamic weighting strategy to adjust the contributions of pseudo-labels using an uncertainty-aware mechanism (i.e., Kullback-Leibler divergence). In addition, we use a self-supervised contrastive learning mechanism to align uncertain voxel features with reliable class prototypes by effectively differentiating between trustworthy and uncertain predictions, thus reducing prediction uncertainty. Extensive experiments are conducted on three 3D segmentation datasets, Left Atrial, NIH Pancreas and BraTS-2019. The proposed approach consistently exhibits superior performance across various settings (e.g., 89.95\% Dice score on left Atrial with 10\% labeled data) compared to the state-of-the-art methods. Furthermore, the usefulness of the proposed modules is further validated via ablation experiments.
Classification based deep learning models for lung cancer and disease using medical images
Jul 02, 2025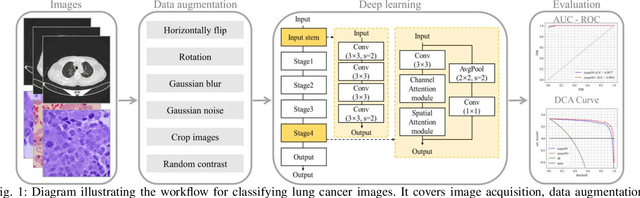
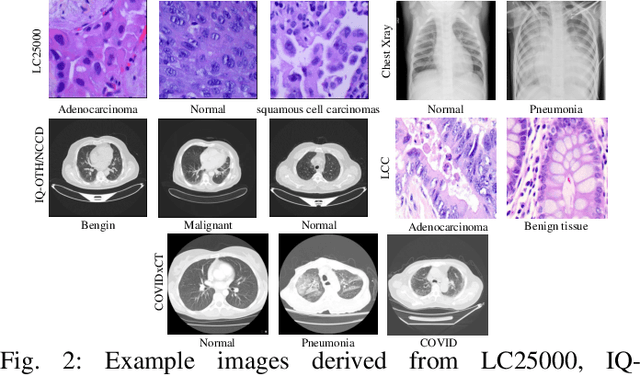
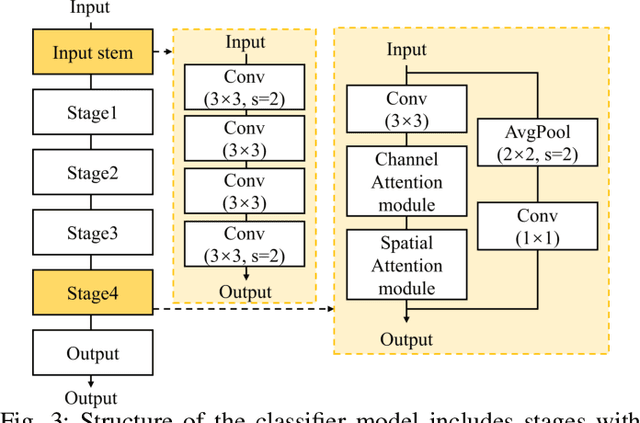
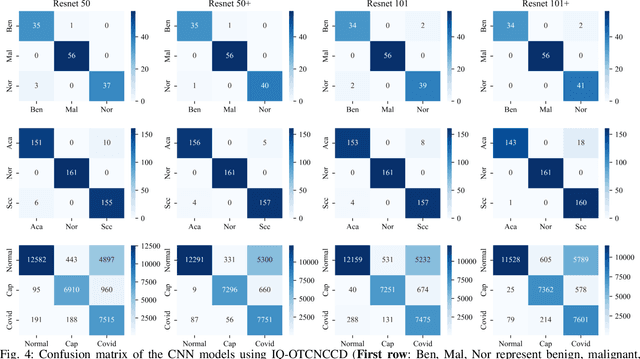
Abstract:The use of deep learning (DL) in medical image analysis has significantly improved the ability to predict lung cancer. In this study, we introduce a novel deep convolutional neural network (CNN) model, named ResNet+, which is based on the established ResNet framework. This model is specifically designed to improve the prediction of lung cancer and diseases using the images. To address the challenge of missing feature information that occurs during the downsampling process in CNNs, we integrate the ResNet-D module, a variant designed to enhance feature extraction capabilities by modifying the downsampling layers, into the traditional ResNet model. Furthermore, a convolutional attention module was incorporated into the bottleneck layers to enhance model generalization by allowing the network to focus on relevant regions of the input images. We evaluated the proposed model using five public datasets, comprising lung cancer (LC2500 $n$=3183, IQ-OTH/NCCD $n$=1336, and LCC $n$=25000 images) and lung disease (ChestXray $n$=5856, and COVIDx-CT $n$=425024 images). To address class imbalance, we used data augmentation techniques to artificially increase the representation of underrepresented classes in the training dataset. The experimental results show that ResNet+ model demonstrated remarkable accuracy/F1, reaching 98.14/98.14\% on the LC25000 dataset and 99.25/99.13\% on the IQ-OTH/NCCD dataset. Furthermore, the ResNet+ model saved computational cost compared to the original ResNet series in predicting lung cancer images. The proposed model outperformed the baseline models on publicly available datasets, achieving better performance metrics. Our codes are publicly available at https://github.com/AIPMLab/Graduation-2024/tree/main/Peng.
Deep Modeling and Optimization of Medical Image Classification
May 29, 2025Abstract:Deep models, such as convolutional neural networks (CNNs) and vision transformer (ViT), demonstrate remarkable performance in image classification. However, those deep models require large data to fine-tune, which is impractical in the medical domain due to the data privacy issue. Furthermore, despite the feasible performance of contrastive language image pre-training (CLIP) in the natural domain, the potential of CLIP has not been fully investigated in the medical field. To face these challenges, we considered three scenarios: 1) we introduce a novel CLIP variant using four CNNs and eight ViTs as image encoders for the classification of brain cancer and skin cancer, 2) we combine 12 deep models with two federated learning techniques to protect data privacy, and 3) we involve traditional machine learning (ML) methods to improve the generalization ability of those deep models in unseen domain data. The experimental results indicate that maxvit shows the highest averaged (AVG) test metrics (AVG = 87.03\%) in HAM10000 dataset with multimodal learning, while convnext\_l demonstrates remarkable test with an F1-score of 83.98\% compared to swin\_b with 81.33\% in FL model. Furthermore, the use of support vector machine (SVM) can improve the overall test metrics with AVG of $\sim 2\%$ for swin transformer series in ISIC2018. Our codes are available at https://github.com/AIPMLab/SkinCancerSimulation.
Evaluating the Effectiveness of Black-Box Prompt Optimization as the Scale of LLMs Continues to Grow
May 13, 2025Abstract:Black-Box prompt optimization methods have emerged as a promising strategy for refining input prompts to better align large language models (LLMs), thereby enhancing their task performance. Although these methods have demonstrated encouraging results, most studies and experiments have primarily focused on smaller-scale models (e.g., 7B, 14B) or earlier versions (e.g., GPT-3.5) of LLMs. As the scale of LLMs continues to increase, such as with DeepSeek V3 (671B), it remains an open question whether these black-box optimization techniques will continue to yield significant performance improvements for models of such scale. In response to this, we select three well-known black-box optimization methods and evaluate them on large-scale LLMs (DeepSeek V3 and Gemini 2.0 Flash) across four NLU and NLG datasets. The results show that these black-box prompt optimization methods offer only limited improvements on these large-scale LLMs. Furthermore, we hypothesize that the scale of the model is the primary factor contributing to the limited benefits observed. To explore this hypothesis, we conducted experiments on LLMs of varying sizes (Qwen 2.5 series, ranging from 7B to 72B) and observed an inverse scaling law, wherein the effectiveness of black-box optimization methods diminished as the model size increased.
IPBench: Benchmarking the Knowledge of Large Language Models in Intellectual Property
Apr 22, 2025



Abstract:Intellectual Property (IP) is a unique domain that integrates technical and legal knowledge, making it inherently complex and knowledge-intensive. As large language models (LLMs) continue to advance, they show great potential for processing IP tasks, enabling more efficient analysis, understanding, and generation of IP-related content. However, existing datasets and benchmarks either focus narrowly on patents or cover limited aspects of the IP field, lacking alignment with real-world scenarios. To bridge this gap, we introduce the first comprehensive IP task taxonomy and a large, diverse bilingual benchmark, IPBench, covering 8 IP mechanisms and 20 tasks. This benchmark is designed to evaluate LLMs in real-world intellectual property applications, encompassing both understanding and generation. We benchmark 16 LLMs, ranging from general-purpose to domain-specific models, and find that even the best-performing model achieves only 75.8% accuracy, revealing substantial room for improvement. Notably, open-source IP and law-oriented models lag behind closed-source general-purpose models. We publicly release all data and code of IPBench and will continue to update it with additional IP-related tasks to better reflect real-world challenges in the intellectual property domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge