Wufeng Xue
Steerable Pyramid Transform Enables Robust Left Ventricle Quantification
Jan 20, 2022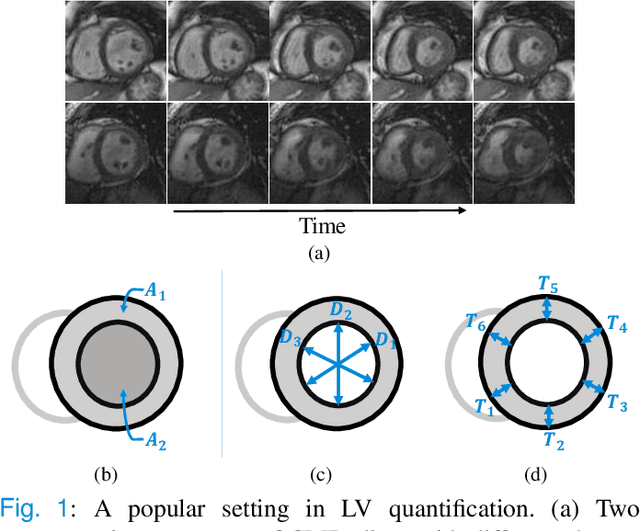
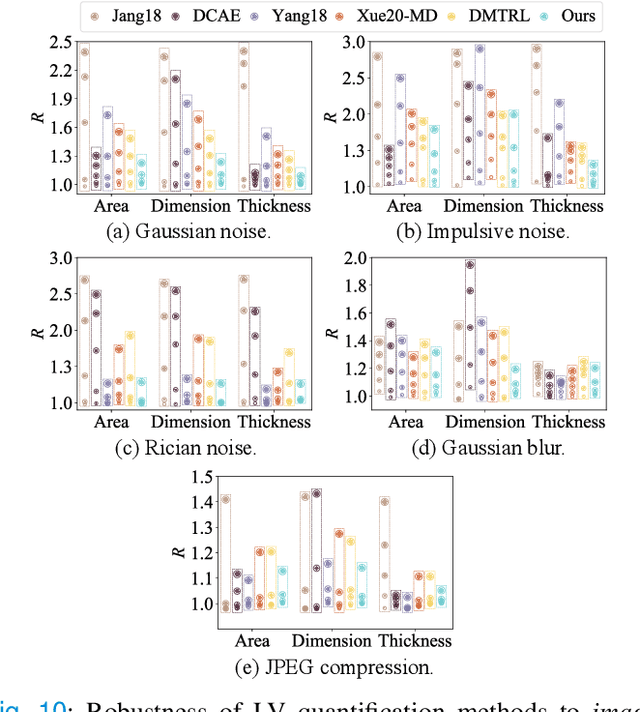
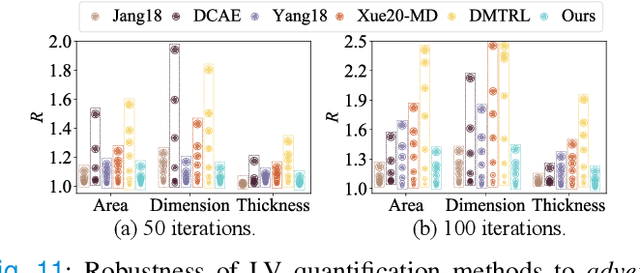
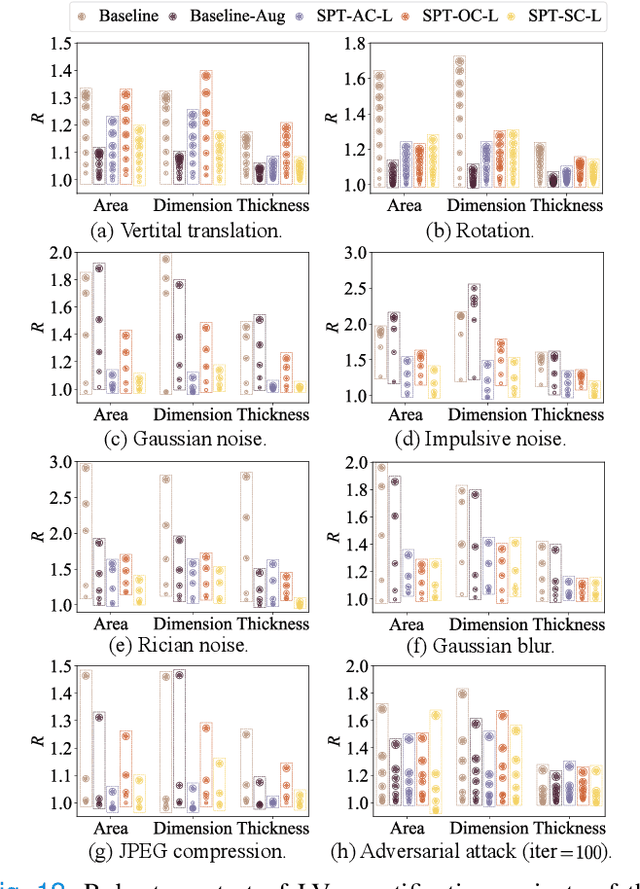
Abstract:Although multifarious variants of convolutional neural networks (CNNs) have proved successful in cardiac index quantification, they seem vulnerable to mild input perturbations, e.g., spatial transformations, image distortions, and adversarial attacks. Such brittleness erodes our trust in CNN-based automated diagnosis of various cardiovascular diseases. In this work, we describe a simple and effective method to learn robust CNNs for left ventricle (LV) quantification, including cavity and myocardium areas, directional dimensions, and regional wall thicknesses. The key to the success of our approach is the use of the biologically-inspired steerable pyramid transform (SPT) as fixed front-end processing, which brings three computational advantages to LV quantification. First, the basis functions of SPT match the anatomical structure of the LV as well as the geometric characteristics of the estimated indices. Second, SPT enables sharing a CNN across different orientations as a form of parameter regularization, and explicitly captures the scale variations of the LV in a natural way. Third, the residual highpass subband can be conveniently discarded to further encourage robust feature learning. A concise and effective metric, named Robustness Ratio, is proposed to evaluate the robustness under various input perturbations. Extensive experiments on 145 cardiac sequences show that our SPT-augmented method performs favorably against state-of-the-art algorithms in terms of prediction accuracy, but is significantly more robust under input perturbations.
AWSnet: An Auto-weighted Supervision Attention Network for Myocardial Scar and Edema Segmentation in Multi-sequence Cardiac Magnetic Resonance Images
Jan 14, 2022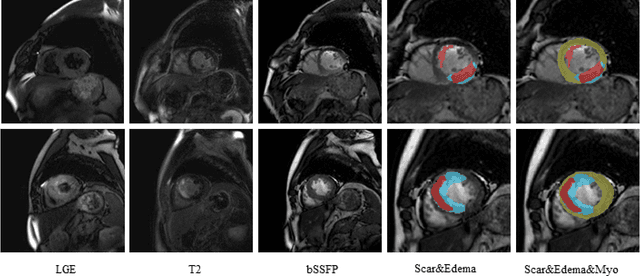
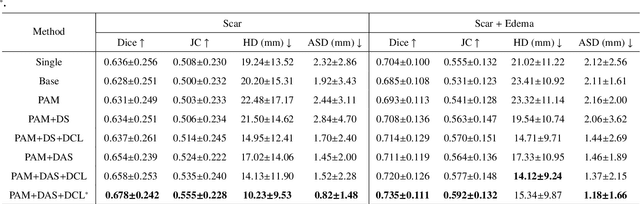
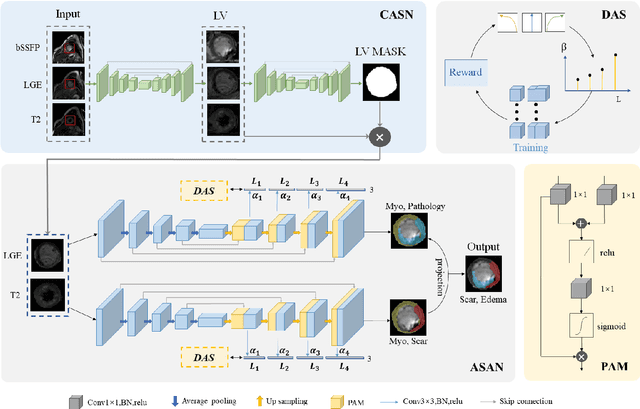
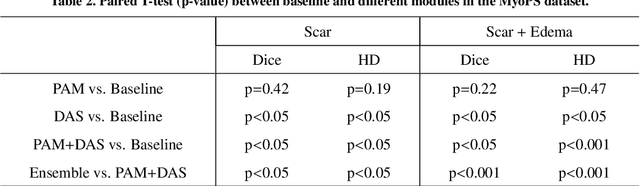
Abstract:Multi-sequence cardiac magnetic resonance (CMR) provides essential pathology information (scar and edema) to diagnose myocardial infarction. However, automatic pathology segmentation can be challenging due to the difficulty of effectively exploring the underlying information from the multi-sequence CMR data. This paper aims to tackle the scar and edema segmentation from multi-sequence CMR with a novel auto-weighted supervision framework, where the interactions among different supervised layers are explored under a task-specific objective using reinforcement learning. Furthermore, we design a coarse-to-fine framework to boost the small myocardial pathology region segmentation with shape prior knowledge. The coarse segmentation model identifies the left ventricle myocardial structure as a shape prior, while the fine segmentation model integrates a pixel-wise attention strategy with an auto-weighted supervision model to learn and extract salient pathological structures from the multi-sequence CMR data. Extensive experimental results on a publicly available dataset from Myocardial pathology segmentation combining multi-sequence CMR (MyoPS 2020) demonstrate our method can achieve promising performance compared with other state-of-the-art methods. Our method is promising in advancing the myocardial pathology assessment on multi-sequence CMR data. To motivate the community, we have made our code publicly available via https://github.com/soleilssss/AWSnet/tree/master.
Bootstrap Representation Learning for Segmentation on Medical Volumes and Sequences
Jun 23, 2021
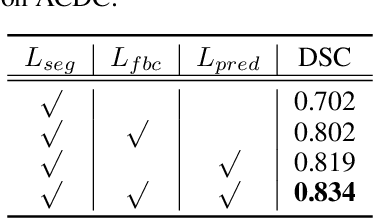
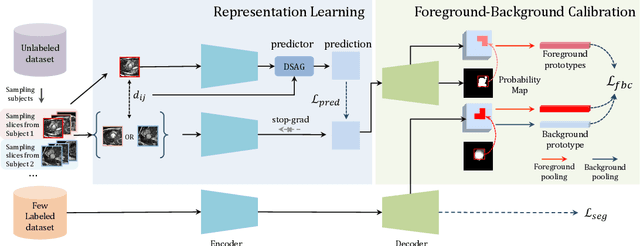
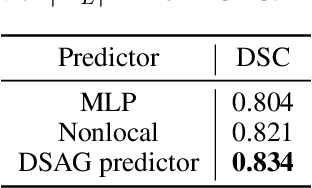
Abstract:In this work, we propose a novel straightforward method for medical volume and sequence segmentation with limited annotations. To avert laborious annotating, the recent success of self-supervised learning(SSL) motivates the pre-training on unlabeled data. Despite its success, it is still challenging to adapt typical SSL methods to volume/sequence segmentation, due to their lack of mining on local semantic discrimination and rare exploitation on volume and sequence structures. Based on the continuity between slices/frames and the common spatial layout of organs across volumes/sequences, we introduced a novel bootstrap self-supervised representation learning method by leveraging the predictable possibility of neighboring slices. At the core of our method is a simple and straightforward dense self-supervision on the predictions of local representations and a strategy of predicting locals based on global context, which enables stable and reliable supervision for both global and local representation mining among volumes. Specifically, we first proposed an asymmetric network with an attention-guided predictor to enforce distance-specific prediction and supervision on slices within and across volumes/sequences. Secondly, we introduced a novel prototype-based foreground-background calibration module to enhance representation consistency. The two parts are trained jointly on labeled and unlabeled data. When evaluated on three benchmark datasets of medical volumes and sequences, our model outperforms existing methods with a large margin of 4.5\% DSC on ACDC, 1.7\% on Prostate, and 2.3\% on CAMUS. Intensive evaluations reveals the effectiveness and superiority of our method.
Joint Landmark and Structure Learning for Automatic Evaluation of Developmental Dysplasia of the Hip
Jun 10, 2021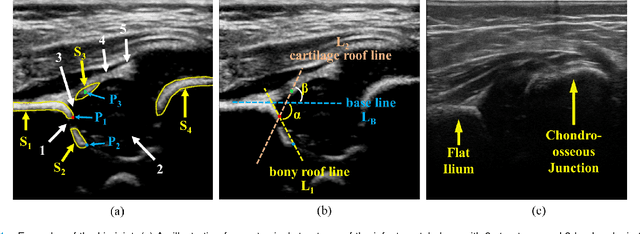
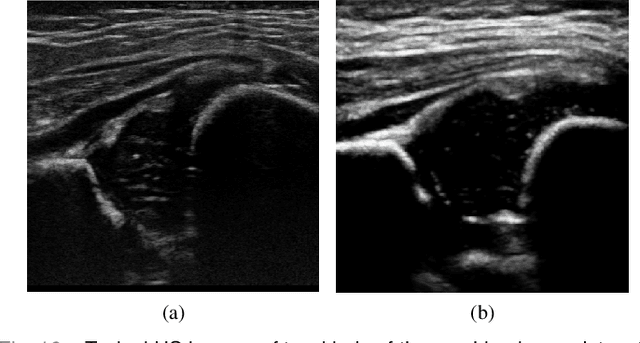
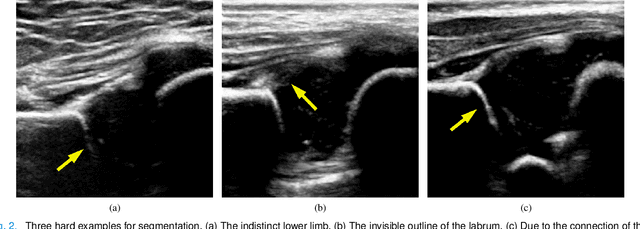
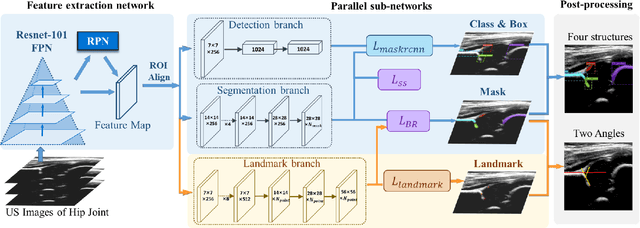
Abstract:The ultrasound (US) screening of the infant hip is vital for the early diagnosis of developmental dysplasia of the hip (DDH). The US diagnosis of DDH refers to measuring alpha and beta angles that quantify hip joint development. These two angles are calculated from key anatomical landmarks and structures of the hip. However, this measurement process is not trivial for sonographers and usually requires a thorough understanding of complex anatomical structures. In this study, we propose a multi-task framework to learn the relationships among landmarks and structures jointly and automatically evaluate DDH. Our multi-task networks are equipped with three novel modules. Firstly, we adopt Mask R-CNN as the basic framework to detect and segment key anatomical structures and add one landmark detection branch to form a new multi-task framework. Secondly, we propose a novel shape similarity loss to refine the incomplete anatomical structure prediction robustly and accurately. Thirdly, we further incorporate the landmark-structure consistent prior to ensure the consistency of the bony rim estimated from the segmented structure and the detected landmark. In our experiments, 1,231 US images of the infant hip from 632 patients are collected, of which 247 images from 126 patients are tested. The average errors in alpha and beta angles are 2.221 degrees and 2.899 degrees. About 93% and 85% estimates of alpha and beta angles have errors less than 5 degrees, respectively. Experimental results demonstrate that the proposed method can accurately and robustly realize the automatic evaluation of DDH, showing great potential for clinical application.
Agent with Warm Start and Adaptive Dynamic Termination for Plane Localization in 3D Ultrasound
Mar 26, 2021
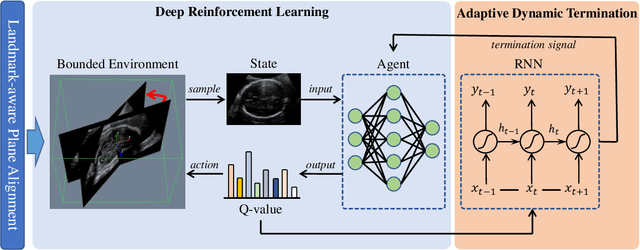
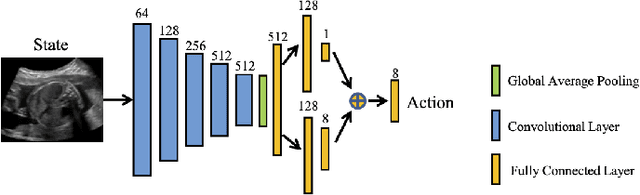
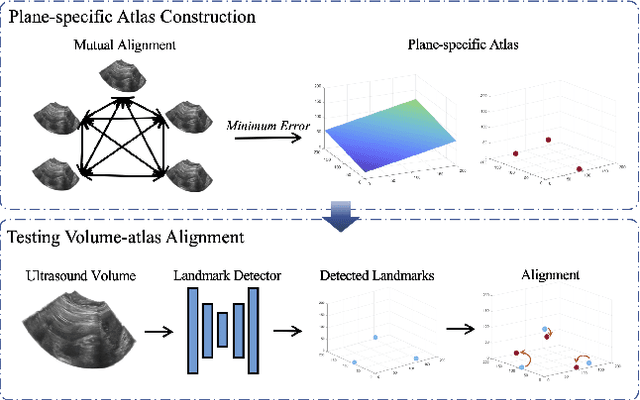
Abstract:Accurate standard plane (SP) localization is the fundamental step for prenatal ultrasound (US) diagnosis. Typically, dozens of US SPs are collected to determine the clinical diagnosis. 2D US has to perform scanning for each SP, which is time-consuming and operator-dependent. While 3D US containing multiple SPs in one shot has the inherent advantages of less user-dependency and more efficiency. Automatically locating SP in 3D US is very challenging due to the huge search space and large fetal posture variations. Our previous study proposed a deep reinforcement learning (RL) framework with an alignment module and active termination to localize SPs in 3D US automatically. However, termination of agent search in RL is important and affects the practical deployment. In this study, we enhance our previous RL framework with a newly designed adaptive dynamic termination to enable an early stop for the agent searching, saving at most 67% inference time, thus boosting the accuracy and efficiency of the RL framework at the same time. Besides, we validate the effectiveness and generalizability of our algorithm extensively on our in-house multi-organ datasets containing 433 fetal brain volumes, 519 fetal abdomen volumes, and 683 uterus volumes. Our approach achieves localization error of 2.52mm/10.26 degrees, 2.48mm/10.39 degrees, 2.02mm/10.48 degrees, 2.00mm/14.57 degrees, 2.61mm/9.71 degrees, 3.09mm/9.58 degrees, 1.49mm/7.54 degrees for the transcerebellar, transventricular, transthalamic planes in fetal brain, abdominal plane in fetal abdomen, and mid-sagittal, transverse and coronal planes in uterus, respectively. Experimental results show that our method is general and has the potential to improve the efficiency and standardization of US scanning.
Style-invariant Cardiac Image Segmentation with Test-time Augmentation
Sep 24, 2020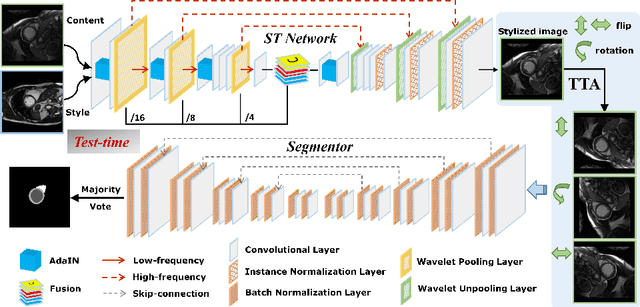
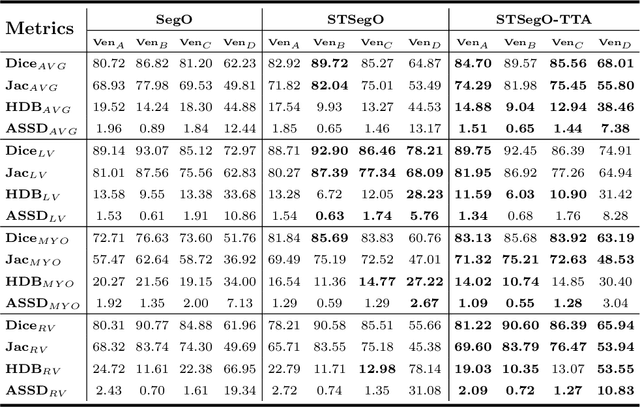
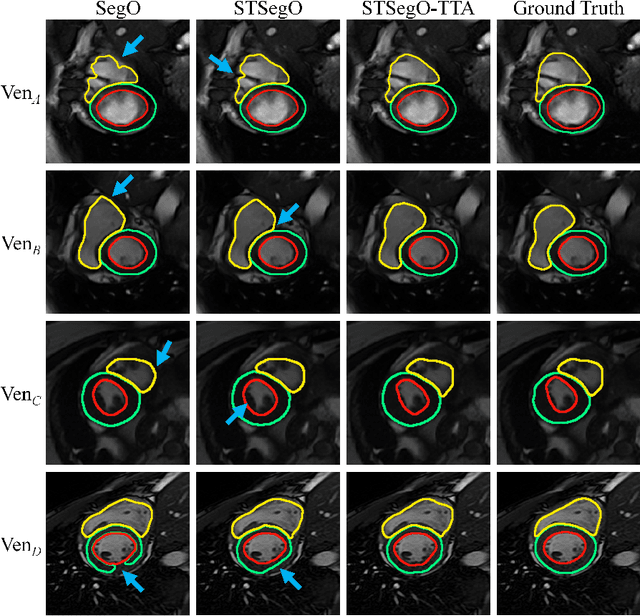
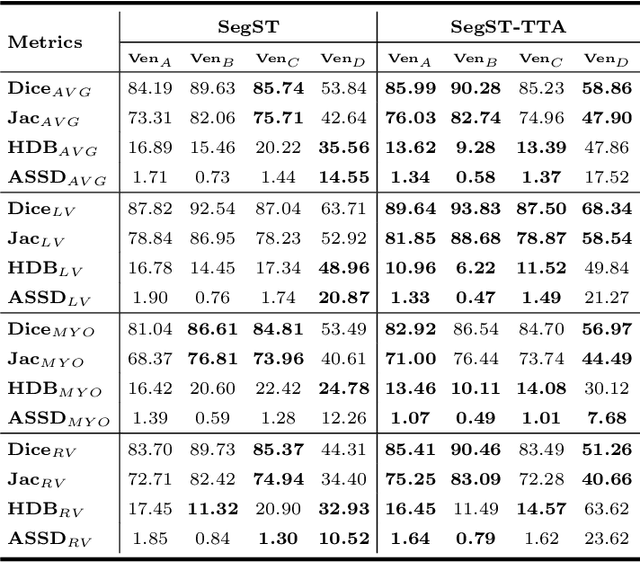
Abstract:Deep models often suffer from severe performance drop due to the appearance shift in the real clinical setting. Most of the existing learning-based methods rely on images from multiple sites/vendors or even corresponding labels. However, collecting enough unknown data to robustly model segmentation cannot always hold since the complex appearance shift caused by imaging factors in daily application. In this paper, we propose a novel style-invariant method for cardiac image segmentation. Based on the zero-shot style transfer to remove appearance shift and test-time augmentation to explore diverse underlying anatomy, our proposed method is effective in combating the appearance shift. Our contribution is three-fold. First, inspired by the spirit of universal style transfer, we develop a zero-shot stylization for content images to generate stylized images that appearance similarity to the style images. Second, we build up a robust cardiac segmentation model based on the U-Net structure. Our framework mainly consists of two networks during testing: the ST network for removing appearance shift and the segmentation network. Third, we investigate test-time augmentation to explore transformed versions of the stylized image for prediction and the results are merged. Notably, our proposed framework is fully test-time adaptation. Experiment results demonstrate that our methods are promising and generic for generalizing deep segmentation models.
FetusMap: Fetal Pose Estimation in 3D Ultrasound
Oct 11, 2019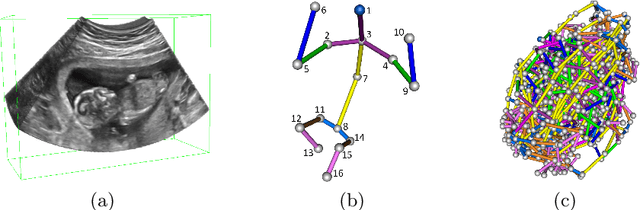
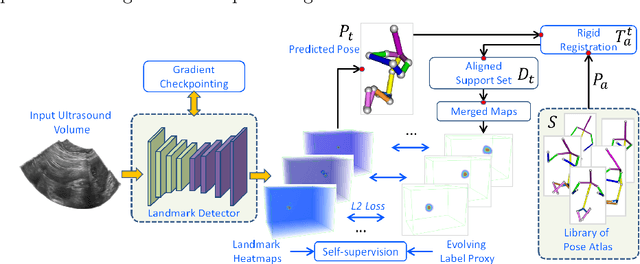
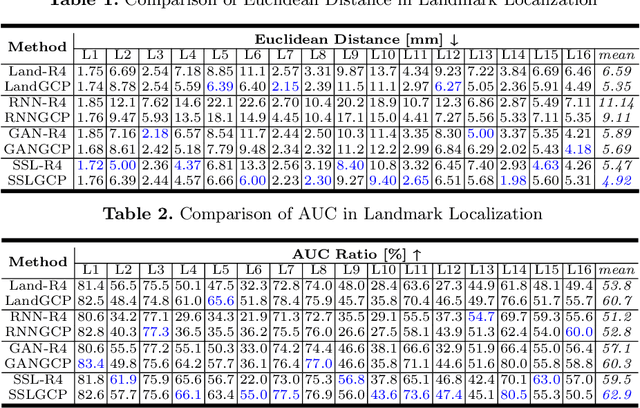
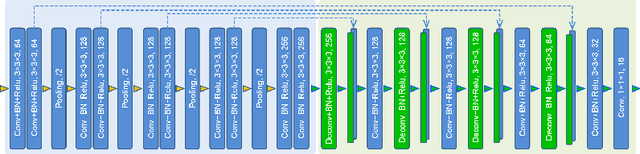
Abstract:The 3D ultrasound (US) entrance inspires a multitude of automated prenatal examinations. However, studies about the structuralized description of the whole fetus in 3D US are still rare. In this paper, we propose to estimate the 3D pose of fetus in US volumes to facilitate its quantitative analyses in global and local scales. Given the great challenges in 3D US, including the high volume dimension, poor image quality, symmetric ambiguity in anatomical structures and large variations of fetal pose, our contribution is three-fold. (i) This is the first work about 3D pose estimation of fetus in the literature. We aim to extract the skeleton of whole fetus and assign different segments/joints with correct torso/limb labels. (ii) We propose a self-supervised learning (SSL) framework to finetune the deep network to form visually plausible pose predictions. Specifically, we leverage the landmark-based registration to effectively encode case-adaptive anatomical priors and generate evolving label proxy for supervision. (iii) To enable our 3D network perceive better contextual cues with higher resolution input under limited computing resource, we further adopt the gradient check-pointing (GCP) strategy to save GPU memory and improve the prediction. Extensively validated on a large 3D US dataset, our method tackles varying fetal poses and achieves promising results. 3D pose estimation of fetus has potentials in serving as a map to provide navigation for many advanced studies.
Agent with Warm Start and Active Termination for Plane Localization in 3D Ultrasound
Oct 10, 2019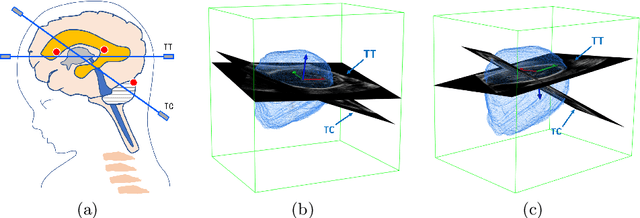
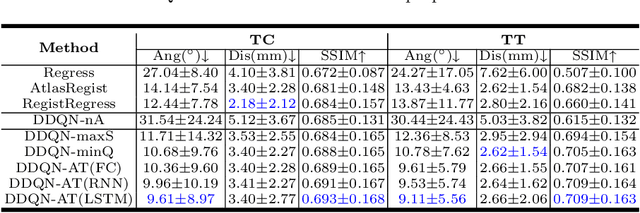
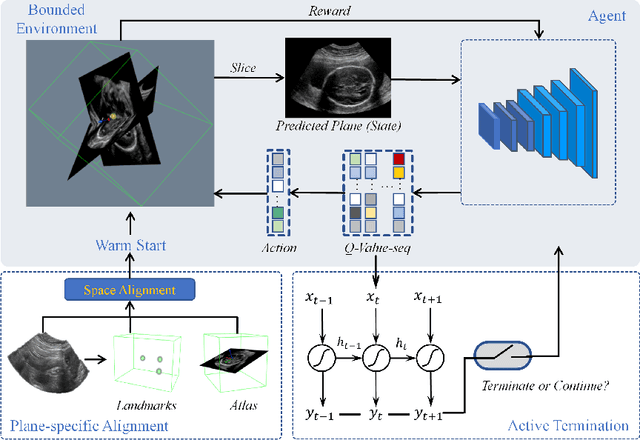
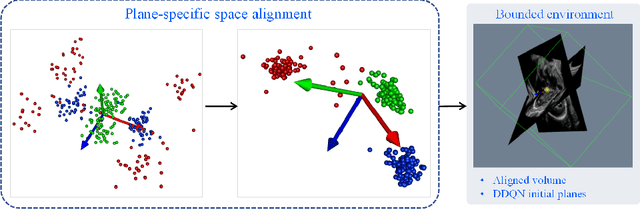
Abstract:Standard plane localization is crucial for ultrasound (US) diagnosis. In prenatal US, dozens of standard planes are manually acquired with a 2D probe. It is time-consuming and operator-dependent. In comparison, 3D US containing multiple standard planes in one shot has the inherent advantages of less user-dependency and more efficiency. However, manual plane localization in US volume is challenging due to the huge search space and large fetal posture variation. In this study, we propose a novel reinforcement learning (RL) framework to automatically localize fetal brain standard planes in 3D US. Our contribution is two-fold. First, we equip the RL framework with a landmark-aware alignment module to provide warm start and strong spatial bounds for the agent actions, thus ensuring its effectiveness. Second, instead of passively and empirically terminating the agent inference, we propose a recurrent neural network based strategy for active termination of the agent's interaction procedure. This improves both the accuracy and efficiency of the localization system. Extensively validated on our in-house large dataset, our approach achieves the accuracy of 3.4mm/9.6{\deg} and 2.7mm/9.1{\deg} for the transcerebellar and transthalamic plane localization, respectively. Ourproposed RL framework is general and has the potential to improve the efficiency and standardization of US scanning.
Segmentation of Multimodal Myocardial Images Using Shape-Transfer GAN
Aug 14, 2019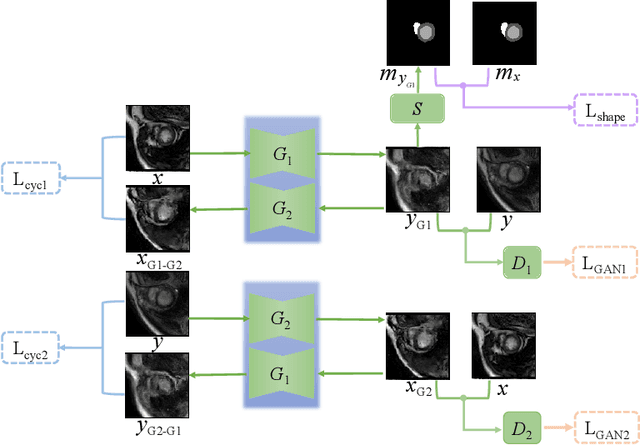
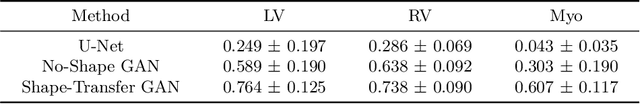
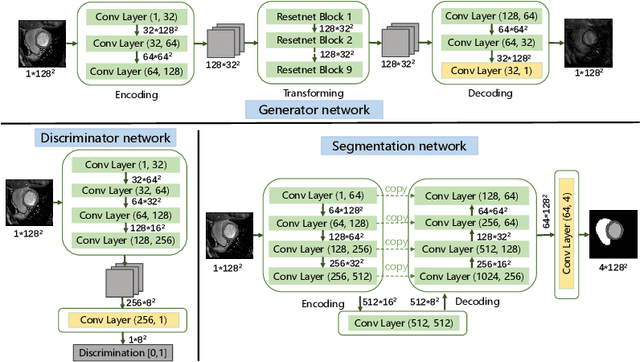
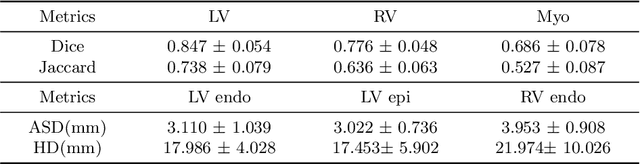
Abstract:Myocardium segmentation of late gadolinium enhancement (LGE) Cardiac MR images is important for evaluation of infarction regions in clinical practice. The pathological myocardium in LGE images presents distinctive brightness and textures compared with the healthy tissues, making it much more challenging to be segment. Instead, the balanced-Steady State Free Precession (bSSFP) cine images show clearly boundaries and can be easily segmented. Given this fact, we propose a novel shape-transfer GAN for LGE images, which can 1) learn to generate realistic LGE images from bSSFP with the anatomical shape preserved, and 2) learn to segment the myocardium of LGE images from these generated images. It's worth to note that no segmentation label of the LGE images is used during this procedure. We test our model on dataset from the Multi-sequence Cardiac MR Segmentation Challenge. The results show that the proposed Shape-Transfer GAN can achieve accurate myocardium masks of LGE images.
Cardiac Motion Scoring with Segment- and Subject-level Non-Local Modeling
Jun 14, 2018
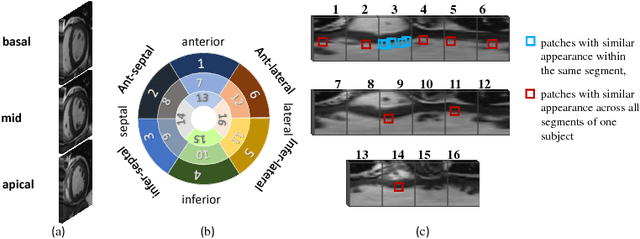
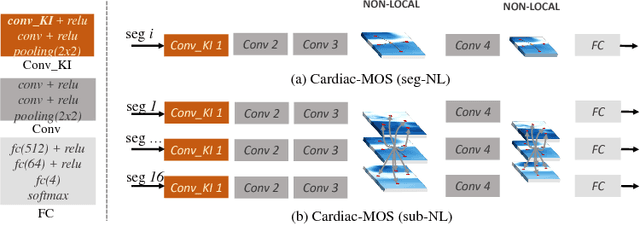
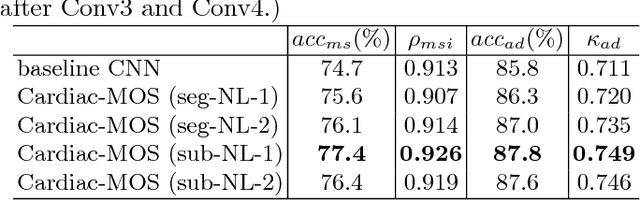
Abstract:Motion scoring of cardiac myocardium is of paramount importance for early detection and diagnosis of various cardiac disease. It aims at identifying regional wall motions into one of the four types including normal, hypokinetic, akinetic, and dyskinetic, and is extremely challenging due to the complex myocardium deformation and subtle inter-class difference of motion patterns. All existing work on automated motion analysis are focused on binary abnormality detection to avoid the much more demanding motion scoring, which is urgently required in real clinical practice yet has never been investigated before. In this work, we propose Cardiac-MOS, the first powerful method for cardiac motion scoring from cardiac MR sequences based on deep convolution neural network. Due to the locality of convolution, the relationship between distant non-local responses of the feature map cannot be explored, which is closely related to motion difference between segments. In Cardiac-MOS, such non-local relationship is modeled with non-local neural network within each segment and across all segments of one subject, i.e., segment- and subject-level non-local modeling, and lead to obvious performance improvement. Besides, Cardiac-MOS can effectively extract motion information from MR sequences of various lengths by interpolating the convolution kernel along the temporal dimension, therefore can be applied to MR sequences of multiple sources. Experiments on 1440 myocardium segments of 90 subjects from short axis MR sequences of multiple lengths prove that Cardiac-MOS achieves reliable performance, with correlation of 0.926 for motion score index estimation and accuracy of 77.4\% for motion scoring. Cardiac-MOS also outperforms all existing work for binary abnormality detection. As the first automatic motion scoring solution, Cardiac-MOS demonstrates great potential in future clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge