Ruobing Huang
Subtyping Breast Lesions via Generative Augmentation based Long-tailed Recognition in Ultrasound
Jul 30, 2025



Abstract:Accurate identification of breast lesion subtypes can facilitate personalized treatment and interventions. Ultrasound (US), as a safe and accessible imaging modality, is extensively employed in breast abnormality screening and diagnosis. However, the incidence of different subtypes exhibits a skewed long-tailed distribution, posing significant challenges for automated recognition. Generative augmentation provides a promising solution to rectify data distribution. Inspired by this, we propose a dual-phase framework for long-tailed classification that mitigates distributional bias through high-fidelity data synthesis while avoiding overuse that corrupts holistic performance. The framework incorporates a reinforcement learning-driven adaptive sampler, dynamically calibrating synthetic-real data ratios by training a strategic multi-agent to compensate for scarcities of real data while ensuring stable discriminative capability. Furthermore, our class-controllable synthetic network integrates a sketch-grounded perception branch that harnesses anatomical priors to maintain distinctive class features while enabling annotation-free inference. Extensive experiments on an in-house long-tailed and a public imbalanced breast US datasets demonstrate that our method achieves promising performance compared to state-of-the-art approaches. More synthetic images can be found at https://github.com/Stinalalala/Breast-LT-GenAug.
Flip Learning: Weakly Supervised Erase to Segment Nodules in Breast Ultrasound
Mar 27, 2025



Abstract:Accurate segmentation of nodules in both 2D breast ultrasound (BUS) and 3D automated breast ultrasound (ABUS) is crucial for clinical diagnosis and treatment planning. Therefore, developing an automated system for nodule segmentation can enhance user independence and expedite clinical analysis. Unlike fully-supervised learning, weakly-supervised segmentation (WSS) can streamline the laborious and intricate annotation process. However, current WSS methods face challenges in achieving precise nodule segmentation, as many of them depend on inaccurate activation maps or inefficient pseudo-mask generation algorithms. In this study, we introduce a novel multi-agent reinforcement learning-based WSS framework called Flip Learning, which relies solely on 2D/3D boxes for accurate segmentation. Specifically, multiple agents are employed to erase the target from the box to facilitate classification tag flipping, with the erased region serving as the predicted segmentation mask. The key contributions of this research are as follows: (1) Adoption of a superpixel/supervoxel-based approach to encode the standardized environment, capturing boundary priors and expediting the learning process. (2) Introduction of three meticulously designed rewards, comprising a classification score reward and two intensity distribution rewards, to steer the agents' erasing process precisely, thereby avoiding both under- and over-segmentation. (3) Implementation of a progressive curriculum learning strategy to enable agents to interact with the environment in a progressively challenging manner, thereby enhancing learning efficiency. Extensively validated on the large in-house BUS and ABUS datasets, our Flip Learning method outperforms state-of-the-art WSS methods and foundation models, and achieves comparable performance as fully-supervised learning algorithms.
EM-Net: Efficient Channel and Frequency Learning with Mamba for 3D Medical Image Segmentation
Sep 26, 2024Abstract:Convolutional neural networks have primarily led 3D medical image segmentation but may be limited by small receptive fields. Transformer models excel in capturing global relationships through self-attention but are challenged by high computational costs at high resolutions. Recently, Mamba, a state space model, has emerged as an effective approach for sequential modeling. Inspired by its success, we introduce a novel Mamba-based 3D medical image segmentation model called EM-Net. It not only efficiently captures attentive interaction between regions by integrating and selecting channels, but also effectively utilizes frequency domain to harmonize the learning of features across varying scales, while accelerating training speed. Comprehensive experiments on two challenging multi-organ datasets with other state-of-the-art (SOTA) algorithms show that our method exhibits better segmentation accuracy while requiring nearly half the parameter size of SOTA models and 2x faster training speed.
Ctrl-GenAug: Controllable Generative Augmentation for Medical Sequence Classification
Sep 25, 2024



Abstract:In the medical field, the limited availability of large-scale datasets and labor-intensive annotation processes hinder the performance of deep models. Diffusion-based generative augmentation approaches present a promising solution to this issue, having been proven effective in advancing downstream medical recognition tasks. Nevertheless, existing works lack sufficient semantic and sequential steerability for challenging video/3D sequence generation, and neglect quality control of noisy synthesized samples, resulting in unreliable synthetic databases and severely limiting the performance of downstream tasks. In this work, we present Ctrl-GenAug, a novel and general generative augmentation framework that enables highly semantic- and sequential-customized sequence synthesis and suppresses incorrectly synthesized samples, to aid medical sequence classification. Specifically, we first design a multimodal conditions-guided sequence generator for controllably synthesizing diagnosis-promotive samples. A sequential augmentation module is integrated to enhance the temporal/stereoscopic coherence of generated samples. Then, we propose a noisy synthetic data filter to suppress unreliable cases at semantic and sequential levels. Extensive experiments on 3 medical datasets, using 11 networks trained on 3 paradigms, comprehensively analyze the effectiveness and generality of Ctrl-GenAug, particularly in underrepresented high-risk populations and out-domain conditions.
TriAug: Out-of-Distribution Detection for Imbalanced Breast Lesion in Ultrasound
Feb 27, 2024



Abstract:Different diseases, such as histological subtypes of breast lesions, have severely varying incidence rates. Even trained with substantial amount of in-distribution (ID) data, models often encounter out-of-distribution (OOD) samples belonging to unseen classes in clinical reality. To address this, we propose a novel framework built upon a long-tailed OOD detection task for breast ultrasound images. It is equipped with a triplet state augmentation (TriAug) which improves ID classification accuracy while maintaining a promising OOD detection performance. Meanwhile, we designed a balanced sphere loss to handle the class imbalanced problem. Experimental results show that the model outperforms state-of-art OOD approaches both in ID classification (F1-score=42.12%) and OOD detection (AUROC=78.06%).
One-Stop Automated Diagnostic System for Carpal Tunnel Syndrome in Ultrasound Images Using Deep Learning
Feb 08, 2024



Abstract:Objective: Ultrasound (US) examination has unique advantages in diagnosing carpal tunnel syndrome (CTS) while identifying the median nerve (MN) and diagnosing CTS depends heavily on the expertise of examiners. To alleviate this problem, we aimed to develop a one-stop automated CTS diagnosis system (OSA-CTSD) and evaluate its effectiveness as a computer-aided diagnostic tool. Methods: We combined real-time MN delineation, accurate biometric measurements, and explainable CTS diagnosis into a unified framework, called OSA-CTSD. We collected a total of 32,301 static images from US videos of 90 normal wrists and 40 CTS wrists for evaluation using a simplified scanning protocol. Results: The proposed model showed better segmentation and measurement performance than competing methods, reporting that HD95 score of 7.21px, ASSD score of 2.64px, Dice score of 85.78%, and IoU score of 76.00%, respectively. In the reader study, it demonstrated comparable performance with the average performance of the experienced in classifying the CTS, while outperformed that of the inexperienced radiologists in terms of classification metrics (e.g., accuracy score of 3.59% higher and F1 score of 5.85% higher). Conclusion: The OSA-CTSD demonstrated promising diagnostic performance with the advantages of real-time, automation, and clinical interpretability. The application of such a tool can not only reduce reliance on the expertise of examiners, but also can help to promote the future standardization of the CTS diagnosis process, benefiting both patients and radiologists.
* Accepted by Ultrasound in Medicine & Biology
Is Two-shot All You Need? A Label-efficient Approach for Video Segmentation in Breast Ultrasound
Feb 07, 2024Abstract:Breast lesion segmentation from breast ultrasound (BUS) videos could assist in early diagnosis and treatment. Existing video object segmentation (VOS) methods usually require dense annotation, which is often inaccessible for medical datasets. Furthermore, they suffer from accumulative errors and a lack of explicit space-time awareness. In this work, we propose a novel two-shot training paradigm for BUS video segmentation. It not only is able to capture free-range space-time consistency but also utilizes a source-dependent augmentation scheme. This label-efficient learning framework is validated on a challenging in-house BUS video dataset. Results showed that it gained comparable performance to the fully annotated ones given only 1.9% training labels.
PE-MED: Prompt Enhancement for Interactive Medical Image Segmentation
Aug 26, 2023



Abstract:Interactive medical image segmentation refers to the accurate segmentation of the target of interest through interaction (e.g., click) between the user and the image. It has been widely studied in recent years as it is less dependent on abundant annotated data and more flexible than fully automated segmentation. However, current studies have not fully explored user-provided prompt information (e.g., points), including the knowledge mined in one interaction, and the relationship between multiple interactions. Thus, in this paper, we introduce a novel framework equipped with prompt enhancement, called PE-MED, for interactive medical image segmentation. First, we introduce a Self-Loop strategy to generate warm initial segmentation results based on the first prompt. It can prevent the highly unfavorable scenarios, such as encountering a blank mask as the initial input after the first interaction. Second, we propose a novel Prompt Attention Learning Module (PALM) to mine useful prompt information in one interaction, enhancing the responsiveness of the network to user clicks. Last, we build a Time Series Information Propagation (TSIP) mechanism to extract the temporal relationships between multiple interactions and increase the model stability. Comparative experiments with other state-of-the-art (SOTA) medical image segmentation algorithms show that our method exhibits better segmentation accuracy and stability.
Hierarchical Agent-based Reinforcement Learning Framework for Automated Quality Assessment of Fetal Ultrasound Video
Apr 14, 2023


Abstract:Ultrasound is the primary modality to examine fetal growth during pregnancy, while the image quality could be affected by various factors. Quality assessment is essential for controlling the quality of ultrasound images to guarantee both the perceptual and diagnostic values. Existing automated approaches often require heavy structural annotations and the predictions may not necessarily be consistent with the assessment results by human experts. Furthermore, the overall quality of a scan and the correlation between the quality of frames should not be overlooked. In this work, we propose a reinforcement learning framework powered by two hierarchical agents that collaboratively learn to perform both frame-level and video-level quality assessments. It is equipped with a specially-designed reward mechanism that considers temporal dependency among frame quality and only requires sparse binary annotations to train. Experimental results on a challenging fetal brain dataset verify that the proposed framework could perform dual-level quality assessment and its predictions correlate well with the subjective assessment results.
Masked Video Modeling with Correlation-aware Contrastive Learning for Breast Cancer Diagnosis in Ultrasound
Aug 21, 2022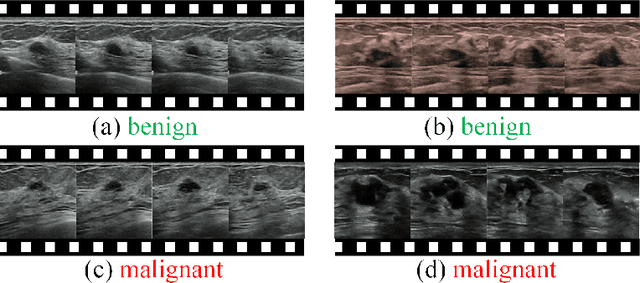
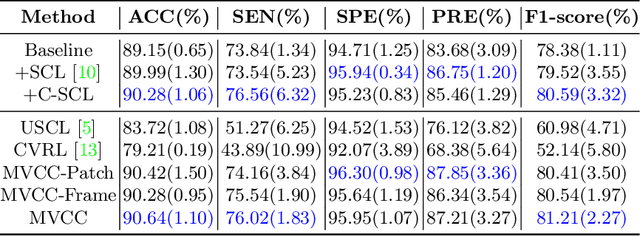
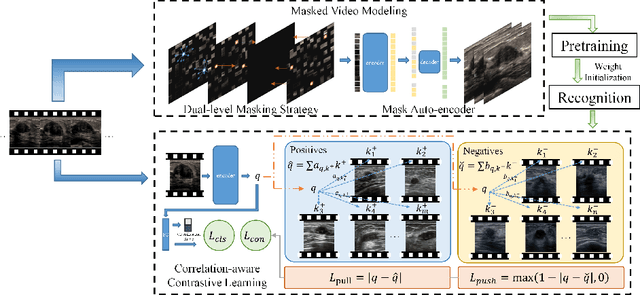
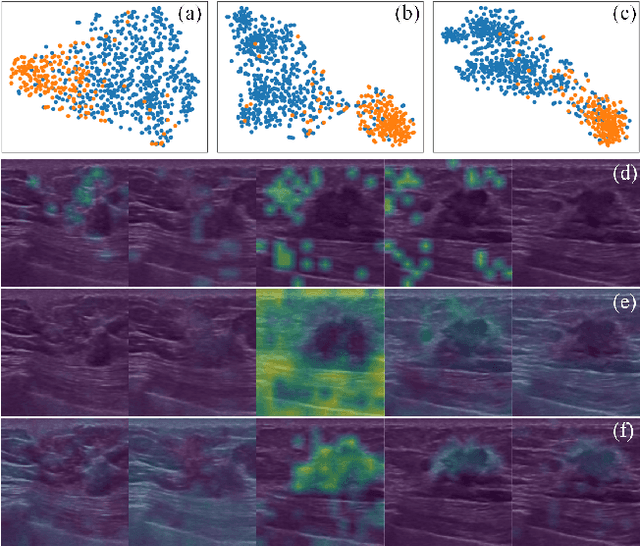
Abstract:Breast cancer is one of the leading causes of cancer deaths in women. As the primary output of breast screening, breast ultrasound (US) video contains exclusive dynamic information for cancer diagnosis. However, training models for video analysis is non-trivial as it requires a voluminous dataset which is also expensive to annotate. Furthermore, the diagnosis of breast lesion faces unique challenges such as inter-class similarity and intra-class variation. In this paper, we propose a pioneering approach that directly utilizes US videos in computer-aided breast cancer diagnosis. It leverages masked video modeling as pretraning to reduce reliance on dataset size and detailed annotations. Moreover, a correlation-aware contrastive loss is developed to facilitate the identifying of the internal and external relationship between benign and malignant lesions. Experimental results show that our proposed approach achieved promising classification performance and can outperform other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge