Ge Liu
Protein Autoregressive Modeling via Multiscale Structure Generation
Feb 04, 2026Abstract:We present protein autoregressive modeling (PAR), the first multi-scale autoregressive framework for protein backbone generation via coarse-to-fine next-scale prediction. Using the hierarchical nature of proteins, PAR generates structures that mimic sculpting a statue, forming a coarse topology and refining structural details over scales. To achieve this, PAR consists of three key components: (i) multi-scale downsampling operations that represent protein structures across multiple scales during training; (ii) an autoregressive transformer that encodes multi-scale information and produces conditional embeddings to guide structure generation; (iii) a flow-based backbone decoder that generates backbone atoms conditioned on these embeddings. Moreover, autoregressive models suffer from exposure bias, caused by the training and the generation procedure mismatch, and substantially degrades structure generation quality. We effectively alleviate this issue by adopting noisy context learning and scheduled sampling, enabling robust backbone generation. Notably, PAR exhibits strong zero-shot generalization, supporting flexible human-prompted conditional generation and motif scaffolding without requiring fine-tuning. On the unconditional generation benchmark, PAR effectively learns protein distributions and produces backbones of high design quality, and exhibits favorable scaling behavior. Together, these properties establish PAR as a promising framework for protein structure generation.
Rethinking the Reranker: Boundary-Aware Evidence Selection for Robust Retrieval-Augmented Generation
Feb 03, 2026Abstract:Retrieval-Augmented Generation (RAG) systems remain brittle under realistic retrieval noise, even when the required evidence appears in the top-K results. A key reason is that retrievers and rerankers optimize solely for relevance, often selecting either trivial, answer-revealing passages or evidence that lacks the critical information required to answer the question, without considering whether the evidence is suitable for the generator. We propose BAR-RAG, which reframes the reranker as a boundary-aware evidence selector that targets the generator's Goldilocks Zone -- evidence that is neither trivially easy nor fundamentally unanswerable for the generator, but is challenging yet sufficient for inference and thus provides the strongest learning signal. BAR-RAG trains the selector with reinforcement learning using generator feedback, and adopts a two-stage pipeline that fine-tunes the generator under the induced evidence distribution to mitigate the distribution mismatch between training and inference. Experiments on knowledge-intensive question answering benchmarks show that BAR-RAG consistently improves end-to-end performance under noisy retrieval, achieving an average gain of 10.3 percent over strong RAG and reranking baselines while substantially improving robustness. Code is publicly avaliable at https://github.com/GasolSun36/BAR-RAG.
Probing the Knowledge Boundary: An Interactive Agentic Framework for Deep Knowledge Extraction
Feb 01, 2026Abstract:Large Language Models (LLMs) can be seen as compressed knowledge bases, but it remains unclear what knowledge they truly contain and how far their knowledge boundaries extend. Existing benchmarks are mostly static and provide limited support for systematic knowledge probing. In this paper, we propose an interactive agentic framework to systematically extract and quantify the knowledge of LLMs. Our method includes four adaptive exploration policies to probe knowledge at different granularities. To ensure the quality of extracted knowledge, we introduce a three-stage knowledge processing pipeline that combines vector-based filtering to remove exact duplicates, LLM-based adjudication to resolve ambiguous semantic overlaps, and domain-relevance auditing to retain valid knowledge units. Through extensive experiments, we find that recursive taxonomy is the most effective exploration strategy. We also observe a clear knowledge scaling law, where larger models consistently extract more knowledge. In addition, we identify a Pass@1-versus-Pass@k trade-off: domain-specialized models achieve higher initial accuracy but degrade rapidly, while general-purpose models maintain stable performance during extended extraction. Finally, our results show that differences in training data composition lead to distinct and measurable knowledge profiles across model families.
UniRec: Unified Multimodal Encoding for LLM-Based Recommendations
Jan 27, 2026Abstract:Large language models have recently shown promise for multimodal recommendation, particularly with text and image inputs. Yet real-world recommendation signals extend far beyond these modalities. To reflect this, we formalize recommendation features into four modalities: text, images, categorical features, and numerical attributes, and highlight the unique challenges this heterogeneity poses for LLMs in understanding multimodal information. In particular, these challenges arise not only across modalities but also within them, as attributes such as price, rating, and time may all be numeric yet carry distinct semantic meanings. Beyond this intra-modality ambiguity, another major challenge is the nested structure of recommendation signals, where user histories are sequences of items, each associated with multiple attributes. To address these challenges, we propose UniRec, a unified multimodal encoder for LLM-based recommendation. UniRec first employs modality-specific encoders to produce consistent embeddings across heterogeneous signals. It then adopts a triplet representation, comprising attribute name, type, and value, to separate schema from raw inputs and preserve semantic distinctions. Finally, a hierarchical Q-Former models the nested structure of user interactions while maintaining their layered organization. Across multiple real-world benchmarks, UniRec outperforms state-of-the-art multimodal and LLM-based recommenders by up to 15%, and extensive ablation studies further validate the contributions of each component.
R$^{2}$Seg: Training-Free OOD Medical Tumor Segmentation via Anatomical Reasoning and Statistical Rejection
Nov 16, 2025Abstract:Foundation models for medical image segmentation struggle under out-of-distribution (OOD) shifts, often producing fragmented false positives on OOD tumors. We introduce R$^{2}$Seg, a training-free framework for robust OOD tumor segmentation that operates via a two-stage Reason-and-Reject process. First, the Reason step employs an LLM-guided anatomical reasoning planner to localize organ anchors and generate multi-scale ROIs. Second, the Reject step applies two-sample statistical testing to candidates generated by a frozen foundation model (BiomedParse) within these ROIs. This statistical rejection filter retains only candidates significantly different from normal tissue, effectively suppressing false positives. Our framework requires no parameter updates, making it compatible with zero-update test-time augmentation and avoiding catastrophic forgetting. On multi-center and multi-modal tumor segmentation benchmarks, R$^{2}$Seg substantially improves Dice, specificity, and sensitivity over strong baselines and the original foundation models. Code are available at https://github.com/Eurekashen/R2Seg.
From Supervision to Exploration: What Does Protein Language Model Learn During Reinforcement Learning?
Oct 02, 2025Abstract:Protein language models (PLMs) have advanced computational protein science through large-scale pretraining and scalable architectures. In parallel, reinforcement learning (RL) has broadened exploration and enabled precise multi-objective optimization in protein design. Yet whether RL can push PLMs beyond their pretraining priors to uncover latent sequence-structure-function rules remains unclear. We address this by pairing RL with PLMs across four domains: antimicrobial peptide design, kinase variant optimization, antibody engineering, and inverse folding. Using diverse RL algorithms and model classes, we ask if RL improves sampling efficiency and, more importantly, if it reveals capabilities not captured by supervised learning. Across benchmarks, RL consistently boosts success rates and sample efficiency. Performance follows a three-factor interaction: task headroom, reward fidelity, and policy capacity jointly determine gains. When rewards are accurate and informative, policies have sufficient capacity, and tasks leave room beyond supervised baselines, improvements scale; when rewards are noisy or capacity is constrained, gains saturate despite exploration. This view yields practical guidance for RL in protein design: prioritize reward modeling and calibration before scaling policy size, match algorithm and regularization strength to task difficulty, and allocate capacity where marginal gains are largest. Implementation is available at https://github.com/chq1155/RL-PLM.
Flow Matching Meets Biology and Life Science: A Survey
Jul 23, 2025Abstract:Over the past decade, advances in generative modeling, such as generative adversarial networks, masked autoencoders, and diffusion models, have significantly transformed biological research and discovery, enabling breakthroughs in molecule design, protein generation, drug discovery, and beyond. At the same time, biological applications have served as valuable testbeds for evaluating the capabilities of generative models. Recently, flow matching has emerged as a powerful and efficient alternative to diffusion-based generative modeling, with growing interest in its application to problems in biology and life sciences. This paper presents the first comprehensive survey of recent developments in flow matching and its applications in biological domains. We begin by systematically reviewing the foundations and variants of flow matching, and then categorize its applications into three major areas: biological sequence modeling, molecule generation and design, and peptide and protein generation. For each, we provide an in-depth review of recent progress. We also summarize commonly used datasets and software tools, and conclude with a discussion of potential future directions. The corresponding curated resources are available at https://github.com/Violet24K/Awesome-Flow-Matching-Meets-Biology.
Variational Supervised Contrastive Learning
Jun 09, 2025Abstract:Contrastive learning has proven to be highly efficient and adaptable in shaping representation spaces across diverse modalities by pulling similar samples together and pushing dissimilar ones apart. However, two key limitations persist: (1) Without explicit regulation of the embedding distribution, semantically related instances can inadvertently be pushed apart unless complementary signals guide pair selection, and (2) excessive reliance on large in-batch negatives and tailored augmentations hinders generalization. To address these limitations, we propose Variational Supervised Contrastive Learning (VarCon), which reformulates supervised contrastive learning as variational inference over latent class variables and maximizes a posterior-weighted evidence lower bound (ELBO) that replaces exhaustive pair-wise comparisons for efficient class-aware matching and grants fine-grained control over intra-class dispersion in the embedding space. Trained exclusively on image data, our experiments on CIFAR-10, CIFAR-100, ImageNet-100, and ImageNet-1K show that VarCon (1) achieves state-of-the-art performance for contrastive learning frameworks, reaching 79.36% Top-1 accuracy on ImageNet-1K and 78.29% on CIFAR-100 with a ResNet-50 encoder while converging in just 200 epochs; (2) yields substantially clearer decision boundaries and semantic organization in the embedding space, as evidenced by KNN classification, hierarchical clustering results, and transfer-learning assessments; and (3) demonstrates superior performance in few-shot learning than supervised baseline and superior robustness across various augmentation strategies.
ProteinZero: Self-Improving Protein Generation via Online Reinforcement Learning
Jun 09, 2025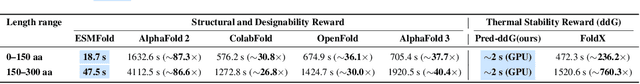
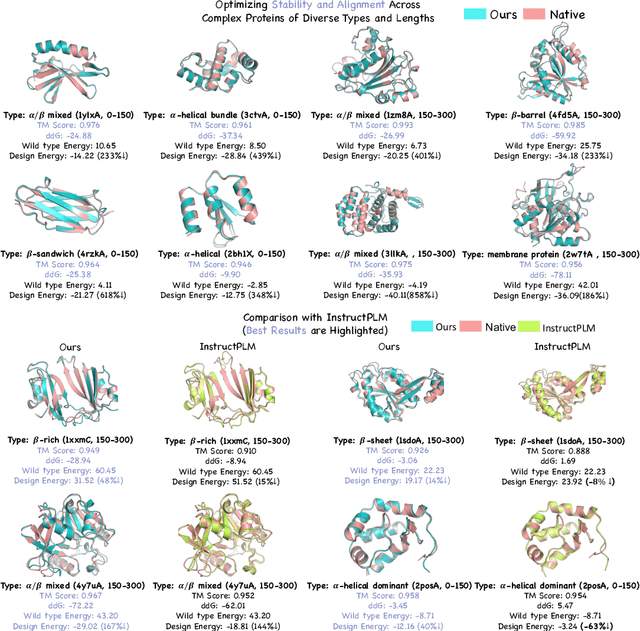
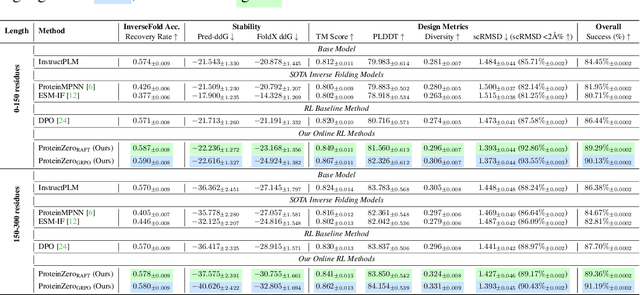
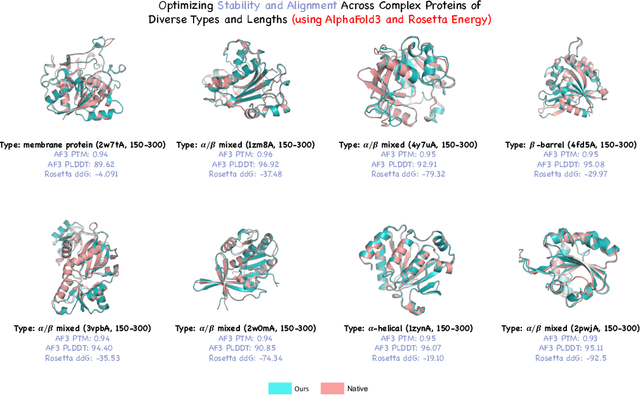
Abstract:Protein generative models have shown remarkable promise in protein design but still face limitations in success rate, due to the scarcity of high-quality protein datasets for supervised pretraining. We present ProteinZero, a novel framework that enables scalable, automated, and continuous self-improvement of the inverse folding model through online reinforcement learning. To achieve computationally tractable online feedback, we introduce efficient proxy reward models based on ESM-fold and a novel rapid ddG predictor that significantly accelerates evaluation speed. ProteinZero employs a general RL framework balancing multi-reward maximization, KL-divergence from a reference model, and a novel protein-embedding level diversity regularization that prevents mode collapse while promoting higher sequence diversity. Through extensive experiments, we demonstrate that ProteinZero substantially outperforms existing methods across every key metric in protein design, achieving significant improvements in structural accuracy, designability, thermodynamic stability, and sequence diversity. Most impressively, ProteinZero reduces design failure rates by approximately 36% - 48% compared to widely-used methods like ProteinMPNN, ESM-IF and InstructPLM, consistently achieving success rates exceeding 90% across diverse and complex protein folds. Notably, the entire RL run on CATH-4.3 can be done with a single 8 X GPU node in under 3 days, including reward computation. Our work establishes a new paradigm for protein design where models evolve continuously from their own generated outputs, opening new possibilities for exploring the vast protein design space.
mCLM: A Function-Infused and Synthesis-Friendly Modular Chemical Language Model
May 18, 2025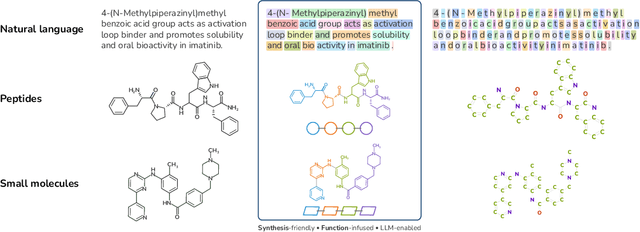
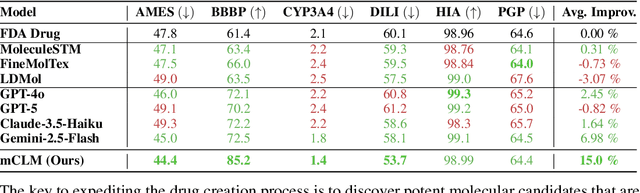
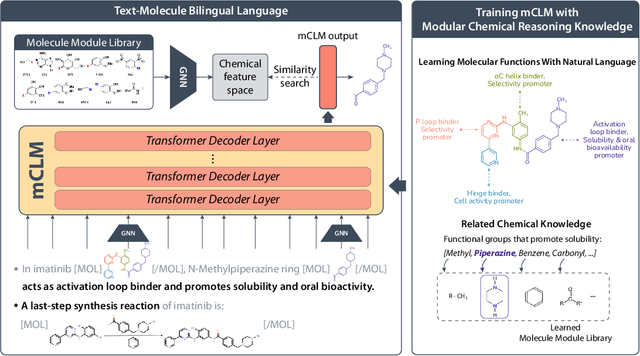
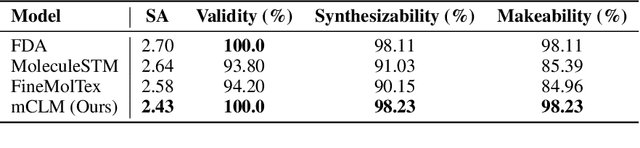
Abstract:Despite their ability to understand chemical knowledge and accurately generate sequential representations, large language models (LLMs) remain limited in their capacity to propose novel molecules with drug-like properties. In addition, the molecules that LLMs propose can often be challenging to make in the lab. To more effectively enable the discovery of functional small molecules, LLMs need to learn a molecular language. However, LLMs are currently limited by encoding molecules from atoms. In this paper, we argue that just like tokenizing texts into (sub-)word tokens instead of characters, molecules should be decomposed and reassembled at the level of functional building blocks, i.e., parts of molecules that bring unique functions and serve as effective building blocks for real-world automated laboratory synthesis. This motivates us to propose mCLM, a modular Chemical-Language Model tokenizing molecules into building blocks and learning a bilingual language model of both natural language descriptions of functions and molecule building blocks. By reasoning on such functional building blocks, mCLM guarantees to generate efficiently synthesizable molecules thanks to recent progress in block-based chemistry, while also improving the functions of molecules in a principled manner. In experiments on 430 FDA-approved drugs, we find mCLM capable of significantly improving 5 out of 6 chemical functions critical to determining drug potentials. More importantly, mCLM can reason on multiple functions and improve the FDA-rejected drugs (``fallen angels'') over multiple iterations to greatly improve their shortcomings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge