Medical Super Resolution
Medical super resolution is the process of enhancing the resolution of medical images to improve their quality.
Papers and Code
Scale-Cascaded Diffusion Models for Super-Resolution in Medical Imaging
Jan 30, 2026Diffusion models have been increasingly used as strong generative priors for solving inverse problems such as super-resolution in medical imaging. However, these approaches typically utilize a diffusion prior trained at a single scale, ignoring the hierarchical scale structure of image data. In this work, we propose to decompose images into Laplacian pyramid scales and train separate diffusion priors for each frequency band. We then develop an algorithm to perform super-resolution that utilizes these priors to progressively refine reconstructions across different scales. Evaluated on brain, knee, and prostate MRI data, our approach both improves perceptual quality over baselines and reduces inference time through smaller coarse-scale networks. Our framework unifies multiscale reconstruction and diffusion priors for medical image super-resolution.
SSPFormer: Self-Supervised Pretrained Transformer for MRI Images
Jan 19, 2026The pre-trained transformer demonstrates remarkable generalization ability in natural image processing. However, directly transferring it to magnetic resonance images faces two key challenges: the inability to adapt to the specificity of medical anatomical structures and the limitations brought about by the privacy and scarcity of medical data. To address these issues, this paper proposes a Self-Supervised Pretrained Transformer (SSPFormer) for MRI images, which effectively learns domain-specific feature representations of medical images by leveraging unlabeled raw imaging data. To tackle the domain gap and data scarcity, we introduce inverse frequency projection masking, which prioritizes the reconstruction of high-frequency anatomical regions to enforce structure-aware representation learning. Simultaneously, to enhance robustness against real-world MRI artifacts, we employ frequency-weighted FFT noise enhancement that injects physiologically realistic noise into the Fourier domain. Together, these strategies enable the model to learn domain-invariant and artifact-robust features directly from raw scans. Through extensive experiments on segmentation, super-resolution, and denoising tasks, the proposed SSPFormer achieves state-of-the-art performance, fully verifying its ability to capture fine-grained MRI image fidelity and adapt to clinical application requirements.
Super-Resolution Enhancement of Medical Images Based on Diffusion Model: An Optimization Scheme for Low-Resolution Gastric Images
Dec 22, 2025Capsule endoscopy has enabled minimally invasive gastrointestinal imaging, but its clinical utility is limited by the inherently low resolution of captured images due to hardware, power, and transmission constraints. This limitation hampers the identification of fine-grained mucosal textures and subtle pathological features essential for early diagnosis. This work investigates a diffusion-based super-resolution framework to enhance capsule endoscopy images in a data-driven and anatomically consistent manner. We adopt the SR3 (Super-Resolution via Repeated Refinement) framework built upon Denoising Diffusion Probabilistic Models (DDPMs) to learn a probabilistic mapping from low-resolution to high-resolution images. Unlike GAN-based approaches that often suffer from training instability and hallucination artifacts, diffusion models provide stable likelihood-based training and improved structural fidelity. The HyperKvasir dataset, a large-scale publicly available gastrointestinal endoscopy dataset, is used for training and evaluation. Quantitative results demonstrate that the proposed method significantly outperforms bicubic interpolation and GAN-based super-resolution methods such as ESRGAN, achieving PSNR of 27.5 dB and SSIM of 0.65 for a baseline model, and improving to 29.3 dB and 0.71 with architectural enhancements including attention mechanisms. Qualitative results show improved preservation of anatomical boundaries, vascular patterns, and lesion structures. These findings indicate that diffusion-based super-resolution is a promising approach for enhancing non-invasive medical imaging, particularly in capsule endoscopy where image resolution is fundamentally constrained.
Fast and Explicit: Slice-to-Volume Reconstruction via 3D Gaussian Primitives with Analytic Point Spread Function Modeling
Dec 16, 2025Recovering high-fidelity 3D images from sparse or degraded 2D images is a fundamental challenge in medical imaging, with broad applications ranging from 3D ultrasound reconstruction to MRI super-resolution. In the context of fetal MRI, high-resolution 3D reconstruction of the brain from motion-corrupted low-resolution 2D acquisitions is a prerequisite for accurate neurodevelopmental diagnosis. While implicit neural representations (INRs) have recently established state-of-the-art performance in self-supervised slice-to-volume reconstruction (SVR), they suffer from a critical computational bottleneck: accurately modeling the image acquisition physics requires expensive stochastic Monte Carlo sampling to approximate the point spread function (PSF). In this work, we propose a shift from neural network based implicit representations to Gaussian based explicit representations. By parameterizing the HR 3D image volume as a field of anisotropic Gaussian primitives, we leverage the property of Gaussians being closed under convolution and thus derive a \textit{closed-form analytical solution} for the forward model. This formulation reduces the previously intractable acquisition integral to an exact covariance addition ($\mathbfΣ_{obs} = \mathbfΣ_{HR} + \mathbfΣ_{PSF}$), effectively bypassing the need for compute-intensive stochastic sampling while ensuring exact gradient propagation. We demonstrate that our approach matches the reconstruction quality of self-supervised state-of-the-art SVR frameworks while delivering a 5$\times$--10$\times$ speed-up on neonatal and fetal data. With convergence often reached in under 30 seconds, our framework paves the way towards translation into clinical routine of real-time fetal 3D MRI. Code will be public at {https://github.com/m-dannecker/Gaussian-Primitives-for-Fast-SVR}.
TAT: Task-Adaptive Transformer for All-in-One Medical Image Restoration
Dec 16, 2025Medical image restoration (MedIR) aims to recover high-quality medical images from their low-quality counterparts. Recent advancements in MedIR have focused on All-in-One models capable of simultaneously addressing multiple different MedIR tasks. However, due to significant differences in both modality and degradation types, using a shared model for these diverse tasks requires careful consideration of two critical inter-task relationships: task interference, which occurs when conflicting gradient update directions arise across tasks on the same parameter, and task imbalance, which refers to uneven optimization caused by varying learning difficulties inherent to each task. To address these challenges, we propose a task-adaptive Transformer (TAT), a novel framework that dynamically adapts to different tasks through two key innovations. First, a task-adaptive weight generation strategy is introduced to mitigate task interference by generating task-specific weight parameters for each task, thereby eliminating potential gradient conflicts on shared weight parameters. Second, a task-adaptive loss balancing strategy is introduced to dynamically adjust loss weights based on task-specific learning difficulties, preventing task domination or undertraining. Extensive experiments demonstrate that our proposed TAT achieves state-of-the-art performance in three MedIR tasks--PET synthesis, CT denoising, and MRI super-resolution--both in task-specific and All-in-One settings. Code is available at https://github.com/Yaziwel/TAT.
Versatile and Efficient Medical Image Super-Resolution Via Frequency-Gated Mamba
Oct 31, 2025Medical image super-resolution (SR) is essential for enhancing diagnostic accuracy while reducing acquisition cost and scanning time. However, modeling both long-range anatomical structures and fine-grained frequency details with low computational overhead remains challenging. We propose FGMamba, a novel frequency-aware gated state-space model that unifies global dependency modeling and fine-detail enhancement into a lightweight architecture. Our method introduces two key innovations: a Gated Attention-enhanced State-Space Module (GASM) that integrates efficient state-space modeling with dual-branch spatial and channel attention, and a Pyramid Frequency Fusion Module (PFFM) that captures high-frequency details across multiple resolutions via FFT-guided fusion. Extensive evaluations across five medical imaging modalities (Ultrasound, OCT, MRI, CT, and Endoscopic) demonstrate that FGMamba achieves superior PSNR/SSIM while maintaining a compact parameter footprint ($<$0.75M), outperforming CNN-based and Transformer-based SOTAs. Our results validate the effectiveness of frequency-aware state-space modeling for scalable and accurate medical image enhancement.
Position-Prior-Guided Network for System Matrix Super-Resolution in Magnetic Particle Imaging
Nov 08, 2025



Magnetic Particle Imaging (MPI) is a novel medical imaging modality. One of the established methods for MPI reconstruction is based on the System Matrix (SM). However, the calibration of the SM is often time-consuming and requires repeated measurements whenever the system parameters change. Current methodologies utilize deep learning-based super-resolution (SR) techniques to expedite SM calibration; nevertheless, these strategies do not fully exploit physical prior knowledge associated with the SM, such as symmetric positional priors. Consequently, we integrated positional priors into existing frameworks for SM calibration. Underpinned by theoretical justification, we empirically validated the efficacy of incorporating positional priors through experiments involving both 2D and 3D SM SR methods.
Rethinking Graph Super-resolution: Dual Frameworks for Topological Fidelity
Nov 12, 2025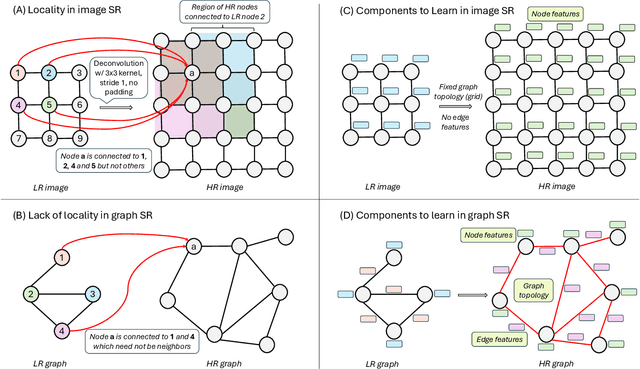
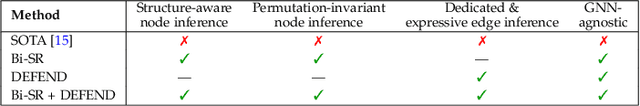
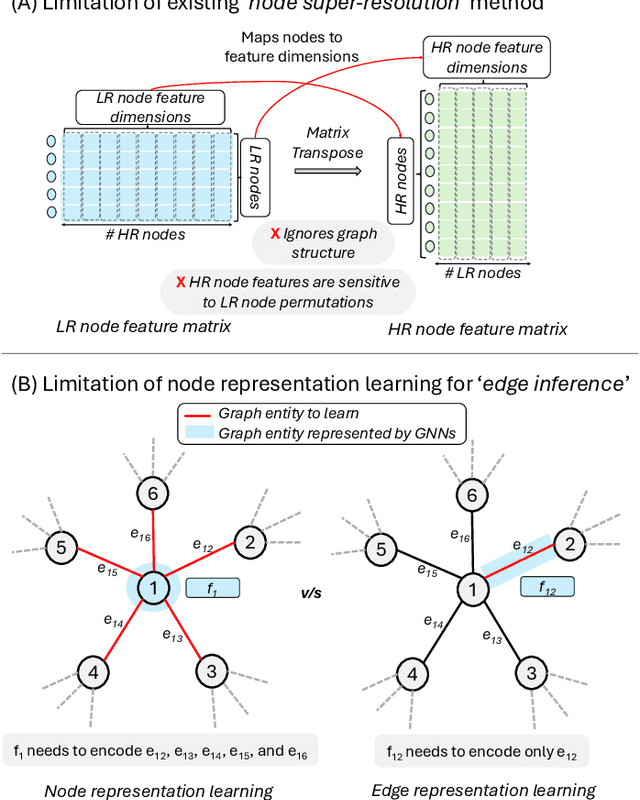
Graph super-resolution, the task of inferring high-resolution (HR) graphs from low-resolution (LR) counterparts, is an underexplored yet crucial research direction that circumvents the need for costly data acquisition. This makes it especially desirable for resource-constrained fields such as the medical domain. While recent GNN-based approaches show promise, they suffer from two key limitations: (1) matrix-based node super-resolution that disregards graph structure and lacks permutation invariance; and (2) reliance on node representations to infer edge weights, which limits scalability and expressivity. In this work, we propose two GNN-agnostic frameworks to address these issues. First, Bi-SR introduces a bipartite graph connecting LR and HR nodes to enable structure-aware node super-resolution that preserves topology and permutation invariance. Second, DEFEND learns edge representations by mapping HR edges to nodes of a dual graph, allowing edge inference via standard node-based GNNs. We evaluate both frameworks on a real-world brain connectome dataset, where they achieve state-of-the-art performance across seven topological measures. To support generalization, we introduce twelve new simulated datasets that capture diverse topologies and LR-HR relationships. These enable comprehensive benchmarking of graph super-resolution methods.
Potential and challenges of generative adversarial networks for super-resolution in 4D Flow MRI
Aug 20, 20254D Flow Magnetic Resonance Imaging (4D Flow MRI) enables non-invasive quantification of blood flow and hemodynamic parameters. However, its clinical application is limited by low spatial resolution and noise, particularly affecting near-wall velocity measurements. Machine learning-based super-resolution has shown promise in addressing these limitations, but challenges remain, not least in recovering near-wall velocities. Generative adversarial networks (GANs) offer a compelling solution, having demonstrated strong capabilities in restoring sharp boundaries in non-medical super-resolution tasks. Yet, their application in 4D Flow MRI remains unexplored, with implementation challenged by known issues such as training instability and non-convergence. In this study, we investigate GAN-based super-resolution in 4D Flow MRI. Training and validation were conducted using patient-specific cerebrovascular in-silico models, converted into synthetic images via an MR-true reconstruction pipeline. A dedicated GAN architecture was implemented and evaluated across three adversarial loss functions: Vanilla, Relativistic, and Wasserstein. Our results demonstrate that the proposed GAN improved near-wall velocity recovery compared to a non-adversarial reference (vNRMSE: 6.9% vs. 9.6%); however, that implementation specifics are critical for stable network training. While Vanilla and Relativistic GANs proved unstable compared to generator-only training (vNRMSE: 8.1% and 7.8% vs. 7.2%), a Wasserstein GAN demonstrated optimal stability and incremental improvement (vNRMSE: 6.9% vs. 7.2%). The Wasserstein GAN further outperformed the generator-only baseline at low SNR (vNRMSE: 8.7% vs. 10.7%). These findings highlight the potential of GAN-based super-resolution in enhancing 4D Flow MRI, particularly in challenging cerebrovascular regions, while emphasizing the need for careful selection of adversarial strategies.
Faster, Self-Supervised Super-Resolution for Anisotropic Multi-View MRI Using a Sparse Coordinate Loss
Sep 09, 2025Acquiring images in high resolution is often a challenging task. Especially in the medical sector, image quality has to be balanced with acquisition time and patient comfort. To strike a compromise between scan time and quality for Magnetic Resonance (MR) imaging, two anisotropic scans with different low-resolution (LR) orientations can be acquired. Typically, LR scans are analyzed individually by radiologists, which is time consuming and can lead to inaccurate interpretation. To tackle this, we propose a novel approach for fusing two orthogonal anisotropic LR MR images to reconstruct anatomical details in a unified representation. Our multi-view neural network is trained in a self-supervised manner, without requiring corresponding high-resolution (HR) data. To optimize the model, we introduce a sparse coordinate-based loss, enabling the integration of LR images with arbitrary scaling. We evaluate our method on MR images from two independent cohorts. Our results demonstrate comparable or even improved super-resolution (SR) performance compared to state-of-the-art (SOTA) self-supervised SR methods for different upsampling scales. By combining a patient-agnostic offline and a patient-specific online phase, we achieve a substantial speed-up of up to ten times for patient-specific reconstruction while achieving similar or better SR quality. Code is available at https://github.com/MajaSchle/tripleSR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge