François Rousseau
IMT Atlantique - ITI, LaTIM
Discovering Data Manifold Geometry via Non-Contracting Flows
Feb 02, 2026Abstract:We introduce an unsupervised approach for constructing a global reference system by learning, in the ambient space, vector fields that span the tangent spaces of an unknown data manifold. In contrast to isometric objectives, which implicitly assume manifold flatness, our method learns tangent vector fields whose flows transport all samples to a common, learnable reference point. The resulting arc-lengths along these flows define interpretable intrinsic coordinates tied to a shared global frame. To prevent degenerate collapse, we enforce a non-shrinking constraint and derive a scalable, integration-free objective inspired by flow matching. Within our theoretical framework, we prove that minimizing the proposed objective recovers a global coordinate chart when one exists. Empirically, we obtain correct tangent alignment and coherent global coordinate structure on synthetic manifolds. We also demonstrate the scalability of our method on CIFAR-10, where the learned coordinates achieve competitive downstream classification performance.
Fast and Explicit: Slice-to-Volume Reconstruction via 3D Gaussian Primitives with Analytic Point Spread Function Modeling
Dec 16, 2025Abstract:Recovering high-fidelity 3D images from sparse or degraded 2D images is a fundamental challenge in medical imaging, with broad applications ranging from 3D ultrasound reconstruction to MRI super-resolution. In the context of fetal MRI, high-resolution 3D reconstruction of the brain from motion-corrupted low-resolution 2D acquisitions is a prerequisite for accurate neurodevelopmental diagnosis. While implicit neural representations (INRs) have recently established state-of-the-art performance in self-supervised slice-to-volume reconstruction (SVR), they suffer from a critical computational bottleneck: accurately modeling the image acquisition physics requires expensive stochastic Monte Carlo sampling to approximate the point spread function (PSF). In this work, we propose a shift from neural network based implicit representations to Gaussian based explicit representations. By parameterizing the HR 3D image volume as a field of anisotropic Gaussian primitives, we leverage the property of Gaussians being closed under convolution and thus derive a \textit{closed-form analytical solution} for the forward model. This formulation reduces the previously intractable acquisition integral to an exact covariance addition ($\mathbfΣ_{obs} = \mathbfΣ_{HR} + \mathbfΣ_{PSF}$), effectively bypassing the need for compute-intensive stochastic sampling while ensuring exact gradient propagation. We demonstrate that our approach matches the reconstruction quality of self-supervised state-of-the-art SVR frameworks while delivering a 5$\times$--10$\times$ speed-up on neonatal and fetal data. With convergence often reached in under 30 seconds, our framework paves the way towards translation into clinical routine of real-time fetal 3D MRI. Code will be public at {https://github.com/m-dannecker/Gaussian-Primitives-for-Fast-SVR}.
Turning Normalizing Flows into Monge Maps with Geodesic Gaussian Preserving Flows
Sep 29, 2022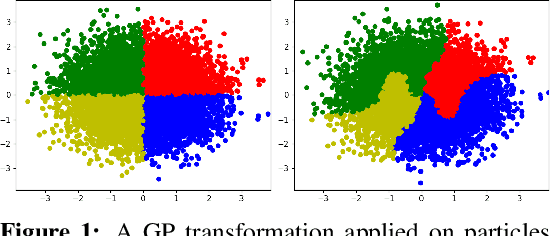
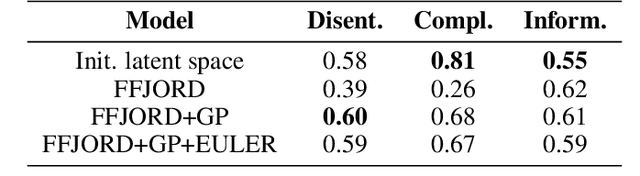
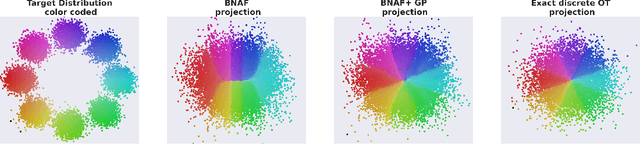
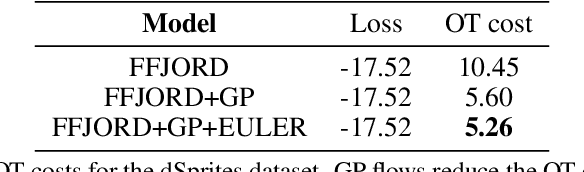
Abstract:Normalizing Flows (NF) are powerful likelihood-based generative models that are able to trade off between expressivity and tractability to model complex densities. A now well established research avenue leverages optimal transport (OT) and looks for Monge maps, i.e. models with minimal effort between the source and target distributions. This paper introduces a method based on Brenier's polar factorization theorem to transform any trained NF into a more OT-efficient version without changing the final density. We do so by learning a rearrangement of the source (Gaussian) distribution that minimizes the OT cost between the source and the final density. We further constrain the path leading to the estimated Monge map to lie on a geodesic in the space of volume-preserving diffeomorphisms thanks to Euler's equations. The proposed method leads to smooth flows with reduced OT cost for several existing models without affecting the model performance.
Abdominal multi-organ segmentation with cascaded convolutional and adversarial deep networks
Jan 26, 2020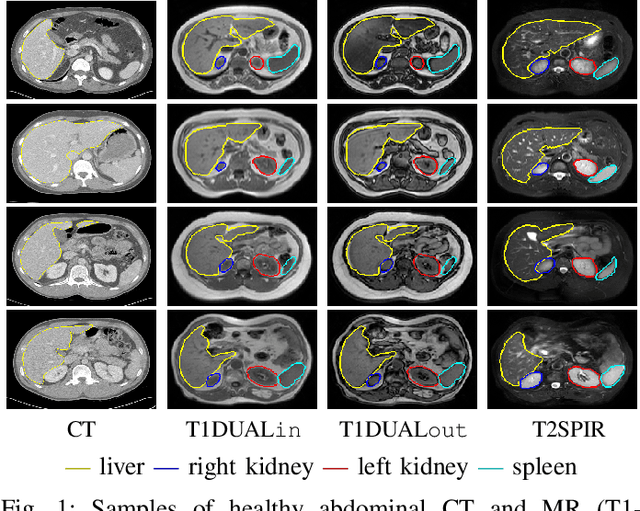
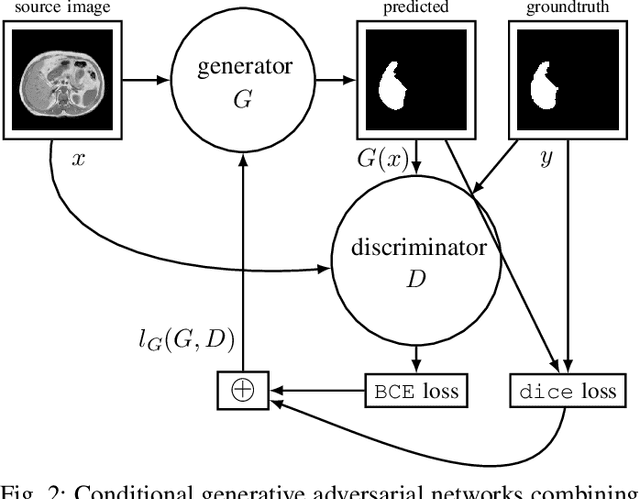
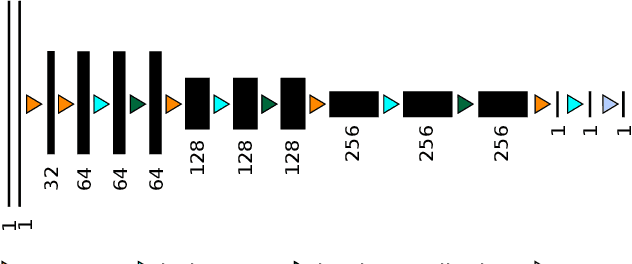
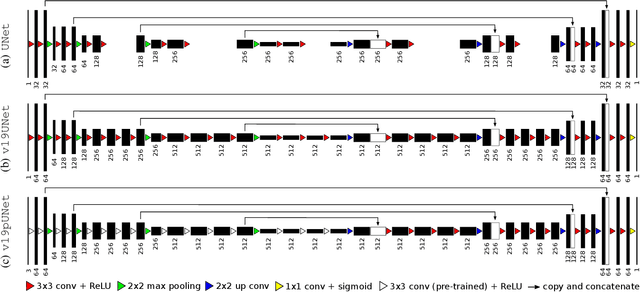
Abstract:Objective : Abdominal anatomy segmentation is crucial for numerous applications from computer-assisted diagnosis to image-guided surgery. In this context, we address fully-automated multi-organ segmentation from abdominal CT and MR images using deep learning. Methods: The proposed model extends standard conditional generative adversarial networks. Additionally to the discriminator which enforces the model to create realistic organ delineations, it embeds cascaded partially pre-trained convolutional encoder-decoders as generator. Encoder fine-tuning from a large amount of non-medical images alleviates data scarcity limitations. The network is trained end-to-end to benefit from simultaneous multi-level segmentation refinements using auto-context. Results : Employed for healthy liver, kidneys and spleen segmentation, our pipeline provides promising results by outperforming state-of-the-art encoder-decoder schemes. Followed for the Combined Healthy Abdominal Organ Segmentation (CHAOS) challenge organized in conjunction with the IEEE International Symposium on Biomedical Imaging 2019, it gave us the first rank for three competition categories: liver CT, liver MR and multi-organ MR segmentation. Conclusion : Combining cascaded convolutional and adversarial networks strengthens the ability of deep learning pipelines to automatically delineate multiple abdominal organs, with good generalization capability. Significance : The comprehensive evaluation provided suggests that better guidance could be achieved to help clinicians in abdominal image interpretation and clinical decision making.
End-to-end learning of energy-based representations for irregularly-sampled signals and images
Oct 01, 2019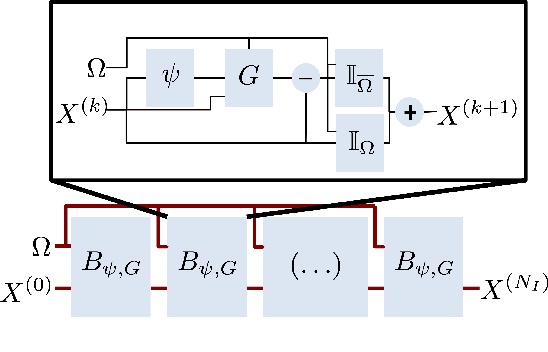
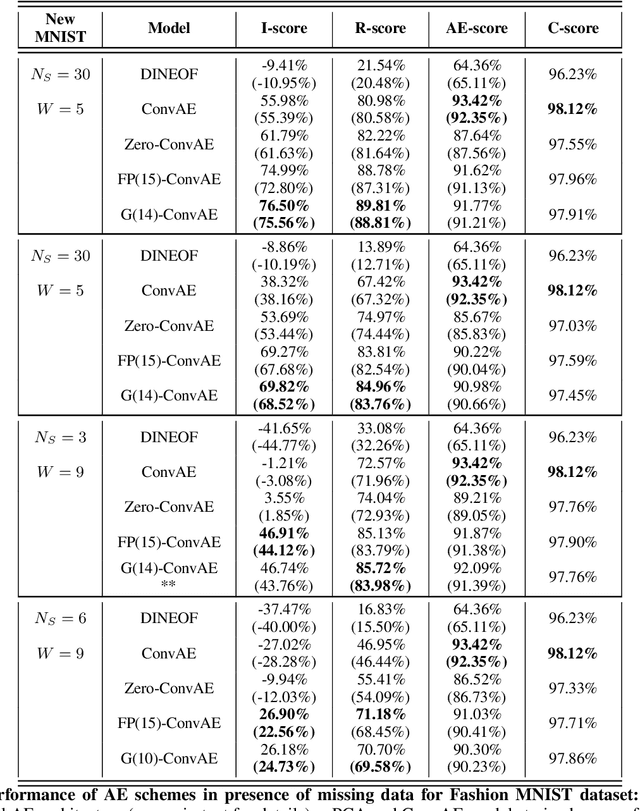
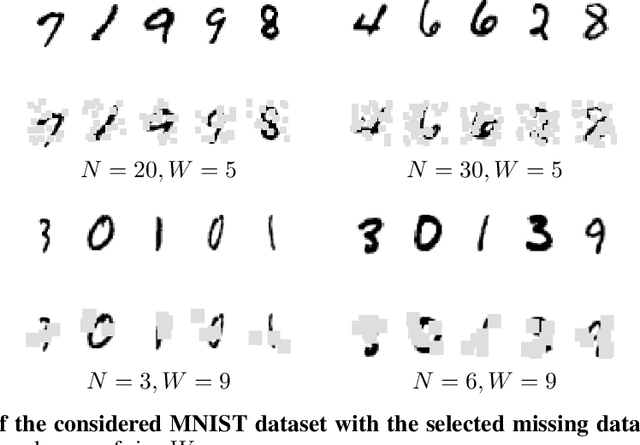
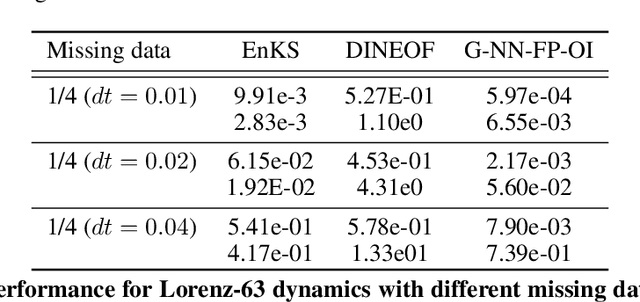
Abstract:For numerous domains, including for instance earth observation, medical imaging, astrophysics,..., available image and signal datasets often involve irregular space-time sampling patterns and large missing data rates. These sampling properties may be critical to apply state-of-the-art learning-based (e.g., auto-encoders, CNNs,...), fully benefit from the available large-scale observations and reach breakthroughs in the reconstruction and identification of processes of interest. In this paper, we address the end-to-end learning of representations of signals, images and image sequences from irregularly-sampled data, i.e. when the training data involved missing data. From an analogy to Bayesian formulation, we consider energy-based representations. Two energy forms are investigated: one derived from auto-encoders and one relating to Gibbs priors. The learning stage of these energy-based representations (or priors) involve a joint interpolation issue, which amounts to solving an energy minimization problem under observation constraints. Using a neural-network-based implementation of the considered energy forms, we can state an end-to-end learning scheme from irregularly-sampled data. We demonstrate the relevance of the proposed representations for different case-studies: namely, multivariate time series, 2D images and image sequences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge