Pragya Singh
Rethinking Graph Super-resolution: Dual Frameworks for Topological Fidelity
Nov 12, 2025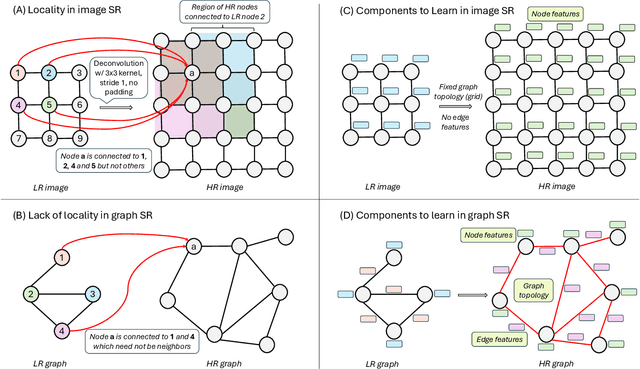
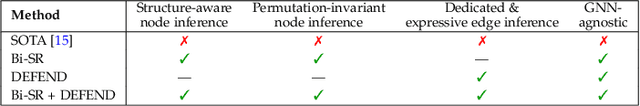
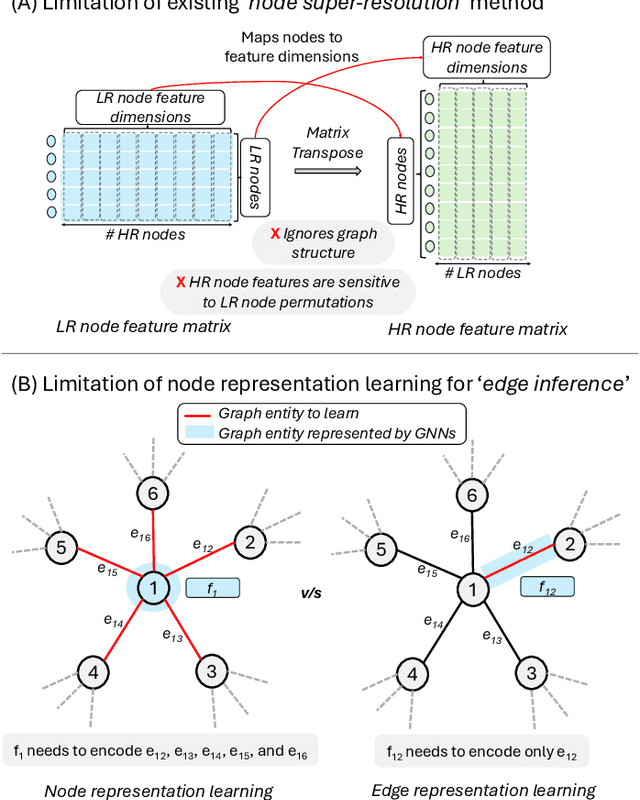
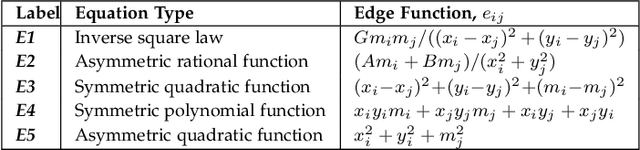
Abstract:Graph super-resolution, the task of inferring high-resolution (HR) graphs from low-resolution (LR) counterparts, is an underexplored yet crucial research direction that circumvents the need for costly data acquisition. This makes it especially desirable for resource-constrained fields such as the medical domain. While recent GNN-based approaches show promise, they suffer from two key limitations: (1) matrix-based node super-resolution that disregards graph structure and lacks permutation invariance; and (2) reliance on node representations to infer edge weights, which limits scalability and expressivity. In this work, we propose two GNN-agnostic frameworks to address these issues. First, Bi-SR introduces a bipartite graph connecting LR and HR nodes to enable structure-aware node super-resolution that preserves topology and permutation invariance. Second, DEFEND learns edge representations by mapping HR edges to nodes of a dual graph, allowing edge inference via standard node-based GNNs. We evaluate both frameworks on a real-world brain connectome dataset, where they achieve state-of-the-art performance across seven topological measures. To support generalization, we introduce twelve new simulated datasets that capture diverse topologies and LR-HR relationships. These enable comprehensive benchmarking of graph super-resolution methods.
Strongly Topology-preserving GNNs for Brain Graph Super-resolution
Nov 01, 2024Abstract:Brain graph super-resolution (SR) is an under-explored yet highly relevant task in network neuroscience. It circumvents the need for costly and time-consuming medical imaging data collection, preparation, and processing. Current SR methods leverage graph neural networks (GNNs) thanks to their ability to natively handle graph-structured datasets. However, most GNNs perform node feature learning, which presents two significant limitations: (1) they require computationally expensive methods to learn complex node features capable of inferring connectivity strength or edge features, which do not scale to larger graphs; and (2) computations in the node space fail to adequately capture higher-order brain topologies such as cliques and hubs. However, numerous studies have shown that brain graph topology is crucial in identifying the onset and presence of various neurodegenerative disorders like Alzheimer and Parkinson. Motivated by these challenges and applications, we propose our STP-GSR framework. It is the first graph SR architecture to perform representation learning in higher-order topological space. Specifically, using the primal-dual graph formulation from graph theory, we develop an efficient mapping from the edge space of our low-resolution (LR) brain graphs to the node space of a high-resolution (HR) dual graph. This approach ensures that node-level computations on this dual graph correspond naturally to edge-level learning on our HR brain graphs, thereby enforcing strong topological consistency within our framework. Additionally, our framework is GNN layer agnostic and can easily learn from smaller, scalable GNNs, reducing computational requirements. We comprehensively benchmark our framework across seven key topological measures and observe that it significantly outperforms the previous state-of-the-art methods and baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge