Tobias Granberg
Potential and challenges of generative adversarial networks for super-resolution in 4D Flow MRI
Aug 20, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) enables non-invasive quantification of blood flow and hemodynamic parameters. However, its clinical application is limited by low spatial resolution and noise, particularly affecting near-wall velocity measurements. Machine learning-based super-resolution has shown promise in addressing these limitations, but challenges remain, not least in recovering near-wall velocities. Generative adversarial networks (GANs) offer a compelling solution, having demonstrated strong capabilities in restoring sharp boundaries in non-medical super-resolution tasks. Yet, their application in 4D Flow MRI remains unexplored, with implementation challenged by known issues such as training instability and non-convergence. In this study, we investigate GAN-based super-resolution in 4D Flow MRI. Training and validation were conducted using patient-specific cerebrovascular in-silico models, converted into synthetic images via an MR-true reconstruction pipeline. A dedicated GAN architecture was implemented and evaluated across three adversarial loss functions: Vanilla, Relativistic, and Wasserstein. Our results demonstrate that the proposed GAN improved near-wall velocity recovery compared to a non-adversarial reference (vNRMSE: 6.9% vs. 9.6%); however, that implementation specifics are critical for stable network training. While Vanilla and Relativistic GANs proved unstable compared to generator-only training (vNRMSE: 8.1% and 7.8% vs. 7.2%), a Wasserstein GAN demonstrated optimal stability and incremental improvement (vNRMSE: 6.9% vs. 7.2%). The Wasserstein GAN further outperformed the generator-only baseline at low SNR (vNRMSE: 8.7% vs. 10.7%). These findings highlight the potential of GAN-based super-resolution in enhancing 4D Flow MRI, particularly in challenging cerebrovascular regions, while emphasizing the need for careful selection of adversarial strategies.
Monitoring morphometric drift in lifelong learning segmentation of the spinal cord
May 02, 2025Abstract:Morphometric measures derived from spinal cord segmentations can serve as diagnostic and prognostic biomarkers in neurological diseases and injuries affecting the spinal cord. While robust, automatic segmentation methods to a wide variety of contrasts and pathologies have been developed over the past few years, whether their predictions are stable as the model is updated using new datasets has not been assessed. This is particularly important for deriving normative values from healthy participants. In this study, we present a spinal cord segmentation model trained on a multisite $(n=75)$ dataset, including 9 different MRI contrasts and several spinal cord pathologies. We also introduce a lifelong learning framework to automatically monitor the morphometric drift as the model is updated using additional datasets. The framework is triggered by an automatic GitHub Actions workflow every time a new model is created, recording the morphometric values derived from the model's predictions over time. As a real-world application of the proposed framework, we employed the spinal cord segmentation model to update a recently-introduced normative database of healthy participants containing commonly used measures of spinal cord morphometry. Results showed that: (i) our model outperforms previous versions and pathology-specific models on challenging lumbar spinal cord cases, achieving an average Dice score of $0.95 \pm 0.03$; (ii) the automatic workflow for monitoring morphometric drift provides a quick feedback loop for developing future segmentation models; and (iii) the scaling factor required to update the database of morphometric measures is nearly constant among slices across the given vertebral levels, showing minimum drift between the current and previous versions of the model monitored by the framework. The model is freely available in Spinal Cord Toolbox v7.0.
Segmentation of Multiple Sclerosis Lesions across Hospitals: Learn Continually or Train from Scratch?
Oct 27, 2022



Abstract:Segmentation of Multiple Sclerosis (MS) lesions is a challenging problem. Several deep-learning-based methods have been proposed in recent years. However, most methods tend to be static, that is, a single model trained on a large, specialized dataset, which does not generalize well. Instead, the model should learn across datasets arriving sequentially from different hospitals by building upon the characteristics of lesions in a continual manner. In this regard, we explore experience replay, a well-known continual learning method, in the context of MS lesion segmentation across multi-contrast data from 8 different hospitals. Our experiments show that replay is able to achieve positive backward transfer and reduce catastrophic forgetting compared to sequential fine-tuning. Furthermore, replay outperforms the multi-domain training, thereby emerging as a promising solution for the segmentation of MS lesions. The code is available at this link: https://github.com/naga-karthik/continual-learning-ms
The reliability of a deep learning model in clinical out-of-distribution MRI data: a multicohort study
Nov 01, 2019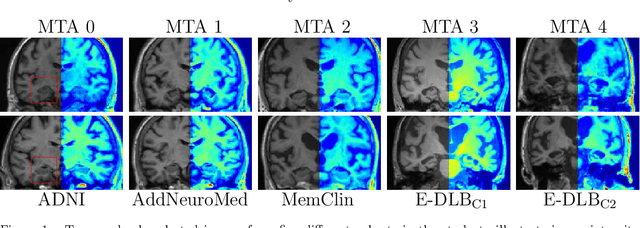
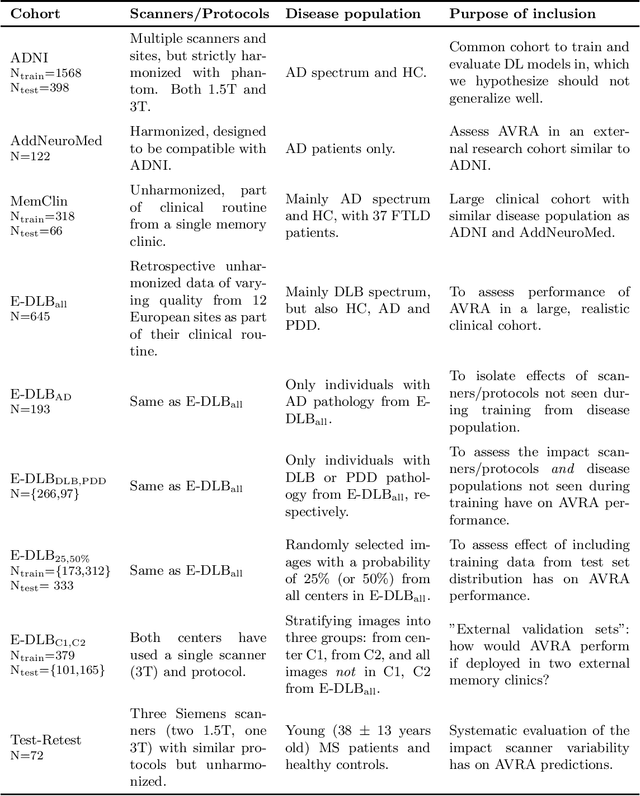
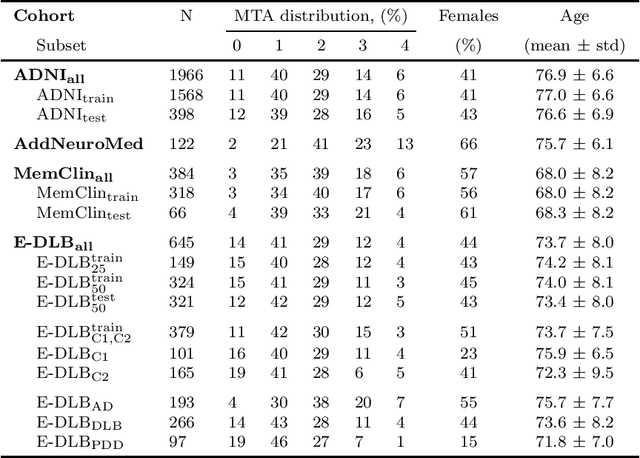
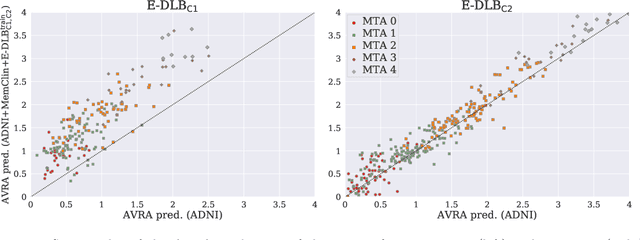
Abstract:Deep learning (DL) methods have in recent years yielded impressive results in medical imaging, with the potential to function as clinical aid to radiologists. However, DL models in medical imaging are often trained on public research cohorts with images acquired with a single scanner or with strict protocol harmonization, which is not representative of a clinical setting. The aim of this study was to investigate how well a DL model performs in unseen clinical data sets---collected with different scanners, protocols and disease populations---and whether more heterogeneous training data improves generalization. In total, 3117 MRI scans of brains from multiple dementia research cohorts and memory clinics, that had been visually rated by a neuroradiologist according to Scheltens' scale of medial temporal atrophy (MTA), were included in this study. By training multiple versions of a convolutional neural network on different subsets of this data to predict MTA ratings, we assessed the impact of including images from a wider distribution during training had on performance in external memory clinic data. Our results showed that our model generalized well to data sets acquired with similar protocols as the training data, but substantially worse in clinical cohorts with visibly different tissue contrasts in the images. This implies that future DL studies investigating performance in out-of-distribution (OOD) MRI data need to assess multiple external cohorts for reliable results. Further, by including data from a wider range of scanners and protocols the performance improved in OOD data, which suggests that more heterogeneous training data makes the model generalize better. To conclude, this is the most comprehensive study to date investigating the domain shift in deep learning on MRI data, and we advocate rigorous evaluation of DL models on clinical data prior to being certified for deployment.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018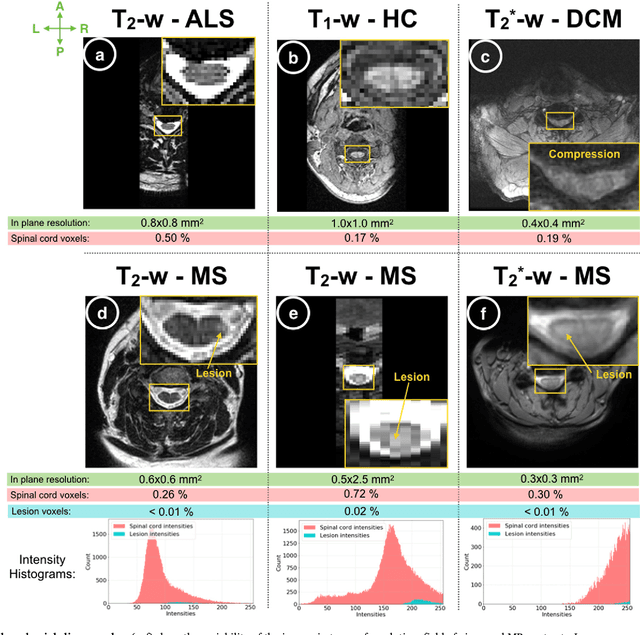
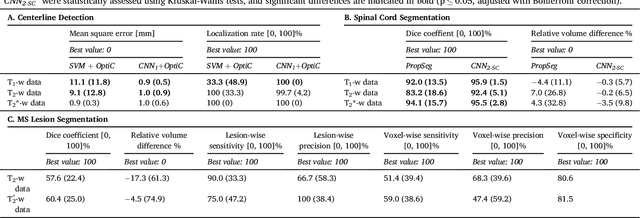
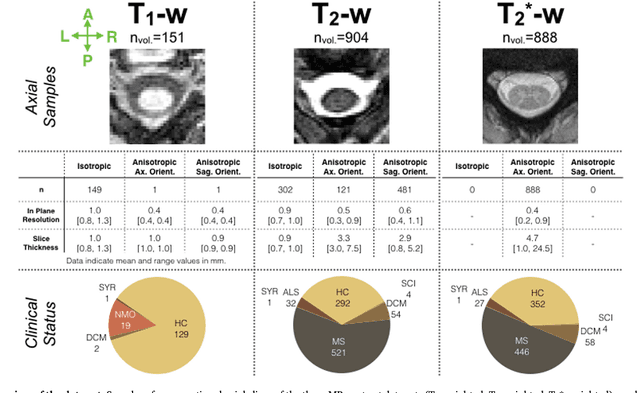
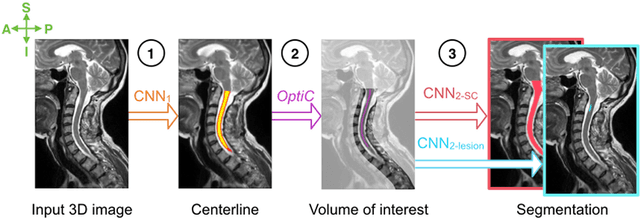
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge