Yan Zhu
GI-Bench: A Panoramic Benchmark Revealing the Knowledge-Experience Dissociation of Multimodal Large Language Models in Gastrointestinal Endoscopy Against Clinical Standards
Jan 13, 2026Abstract:Multimodal Large Language Models (MLLMs) show promise in gastroenterology, yet their performance against comprehensive clinical workflows and human benchmarks remains unverified. To systematically evaluate state-of-the-art MLLMs across a panoramic gastrointestinal endoscopy workflow and determine their clinical utility compared with human endoscopists. We constructed GI-Bench, a benchmark encompassing 20 fine-grained lesion categories. Twelve MLLMs were evaluated across a five-stage clinical workflow: anatomical localization, lesion identification, diagnosis, findings description, and management. Model performance was benchmarked against three junior endoscopists and three residency trainees using Macro-F1, mean Intersection-over-Union (mIoU), and multi-dimensional Likert scale. Gemini-3-Pro achieved state-of-the-art performance. In diagnostic reasoning, top-tier models (Macro-F1 0.641) outperformed trainees (0.492) and rivaled junior endoscopists (0.727; p>0.05). However, a critical "spatial grounding bottleneck" persisted; human lesion localization (mIoU >0.506) significantly outperformed the best model (0.345; p<0.05). Furthermore, qualitative analysis revealed a "fluency-accuracy paradox": models generated reports with superior linguistic readability compared with humans (p<0.05) but exhibited significantly lower factual correctness (p<0.05) due to "over-interpretation" and hallucination of visual features.GI-Bench maintains a dynamic leaderboard that tracks the evolving performance of MLLMs in clinical endoscopy. The current rankings and benchmark results are available at https://roterdl.github.io/GIBench/.
TEA: Temporal Adaptive Satellite Image Semantic Segmentation
Jan 08, 2026Abstract:Crop mapping based on satellite images time-series (SITS) holds substantial economic value in agricultural production settings, in which parcel segmentation is an essential step. Existing approaches have achieved notable advancements in SITS segmentation with predetermined sequence lengths. However, we found that these approaches overlooked the generalization capability of models across scenarios with varying temporal length, leading to markedly poor segmentation results in such cases. To address this issue, we propose TEA, a TEmporal Adaptive SITS semantic segmentation method to enhance the model's resilience under varying sequence lengths. We introduce a teacher model that encapsulates the global sequence knowledge to guide a student model with adaptive temporal input lengths. Specifically, teacher shapes the student's feature space via intermediate embedding, prototypes and soft label perspectives to realize knowledge transfer, while dynamically aggregating student model to mitigate knowledge forgetting. Finally, we introduce full-sequence reconstruction as an auxiliary task to further enhance the quality of representations across inputs of varying temporal lengths. Through extensive experiments, we demonstrate that our method brings remarkable improvements across inputs of different temporal lengths on common benchmarks. Our code will be publicly available.
One-shot synthesis of rare gastrointestinal lesions improves diagnostic accuracy and clinical training
Dec 30, 2025Abstract:Rare gastrointestinal lesions are infrequently encountered in routine endoscopy, restricting the data available for developing reliable artificial intelligence (AI) models and training novice clinicians. Here we present EndoRare, a one-shot, retraining-free generative framework that synthesizes diverse, high-fidelity lesion exemplars from a single reference image. By leveraging language-guided concept disentanglement, EndoRare separates pathognomonic lesion features from non-diagnostic attributes, encoding the former into a learnable prototype embedding while varying the latter to ensure diversity. We validated the framework across four rare pathologies (calcifying fibrous tumor, juvenile polyposis syndrome, familial adenomatous polyposis, and Peutz-Jeghers syndrome). Synthetic images were judged clinically plausible by experts and, when used for data augmentation, significantly enhanced downstream AI classifiers, improving the true positive rate at low false-positive rates. Crucially, a blinded reader study demonstrated that novice endoscopists exposed to EndoRare-generated cases achieved a 0.400 increase in recall and a 0.267 increase in precision. These results establish a practical, data-efficient pathway to bridge the rare-disease gap in both computer-aided diagnostics and clinical education.
Artificial intelligence for representing and characterizing quantum systems
Sep 05, 2025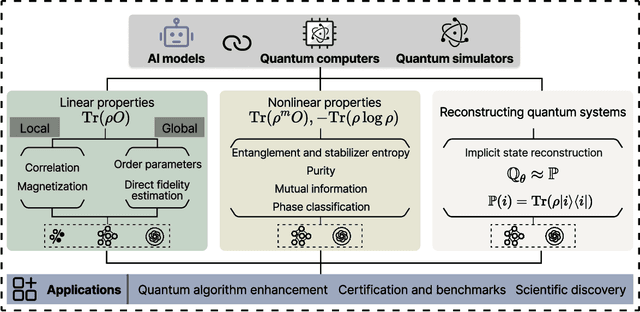

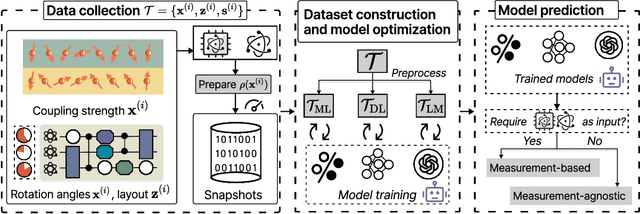
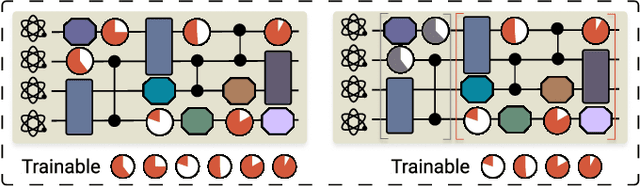
Abstract:Efficient characterization of large-scale quantum systems, especially those produced by quantum analog simulators and megaquop quantum computers, poses a central challenge in quantum science due to the exponential scaling of the Hilbert space with respect to system size. Recent advances in artificial intelligence (AI), with its aptitude for high-dimensional pattern recognition and function approximation, have emerged as a powerful tool to address this challenge. A growing body of research has leveraged AI to represent and characterize scalable quantum systems, spanning from theoretical foundations to experimental realizations. Depending on how prior knowledge and learning architectures are incorporated, the integration of AI into quantum system characterization can be categorized into three synergistic paradigms: machine learning, and, in particular, deep learning and language models. This review discusses how each of these AI paradigms contributes to two core tasks in quantum systems characterization: quantum property prediction and the construction of surrogates for quantum states. These tasks underlie diverse applications, from quantum certification and benchmarking to the enhancement of quantum algorithms and the understanding of strongly correlated phases of matter. Key challenges and open questions are also discussed, together with future prospects at the interface of AI and quantum science.
EndoFinder: Online Lesion Retrieval for Explainable Colorectal Polyp Diagnosis Leveraging Latent Scene Representations
Jul 23, 2025Abstract:Colorectal cancer (CRC) remains a leading cause of cancer-related mortality, underscoring the importance of timely polyp detection and diagnosis. While deep learning models have improved optical-assisted diagnostics, they often demand extensive labeled datasets and yield "black-box" outputs with limited interpretability. In this paper, we propose EndoFinder, an online polyp retrieval framework that leverages multi-view scene representations for explainable and scalable CRC diagnosis. First, we develop a Polyp-aware Image Encoder by combining contrastive learning and a reconstruction task, guided by polyp segmentation masks. This self-supervised approach captures robust features without relying on large-scale annotated data. Next, we treat each polyp as a three-dimensional "scene" and introduce a Scene Representation Transformer, which fuses multiple views of the polyp into a single latent representation. By discretizing this representation through a hashing layer, EndoFinder enables real-time retrieval from a compiled database of historical polyp cases, where diagnostic information serves as interpretable references for new queries. We evaluate EndoFinder on both public and newly collected polyp datasets for re-identification and pathology classification. Results show that EndoFinder outperforms existing methods in accuracy while providing transparent, retrieval-based insights for clinical decision-making. By contributing a novel dataset and a scalable, explainable framework, our work addresses key challenges in polyp diagnosis and offers a promising direction for more efficient AI-driven colonoscopy workflows. The source code is available at https://github.com/ku262/EndoFinder-Scene.
Endo-CLIP: Progressive Self-Supervised Pre-training on Raw Colonoscopy Records
May 14, 2025



Abstract:Pre-training on image-text colonoscopy records offers substantial potential for improving endoscopic image analysis, but faces challenges including non-informative background images, complex medical terminology, and ambiguous multi-lesion descriptions. We introduce Endo-CLIP, a novel self-supervised framework that enhances Contrastive Language-Image Pre-training (CLIP) for this domain. Endo-CLIP's three-stage framework--cleansing, attunement, and unification--addresses these challenges by (1) removing background frames, (2) leveraging large language models to extract clinical attributes for fine-grained contrastive learning, and (3) employing patient-level cross-attention to resolve multi-polyp ambiguities. Extensive experiments demonstrate that Endo-CLIP significantly outperforms state-of-the-art pre-training methods in zero-shot and few-shot polyp detection and classification, paving the way for more accurate and clinically relevant endoscopic analysis.
Robust Polyp Detection and Diagnosis through Compositional Prompt-Guided Diffusion Models
Feb 25, 2025Abstract:Colorectal cancer (CRC) is a significant global health concern, and early detection through screening plays a critical role in reducing mortality. While deep learning models have shown promise in improving polyp detection, classification, and segmentation, their generalization across diverse clinical environments, particularly with out-of-distribution (OOD) data, remains a challenge. Multi-center datasets like PolypGen have been developed to address these issues, but their collection is costly and time-consuming. Traditional data augmentation techniques provide limited variability, failing to capture the complexity of medical images. Diffusion models have emerged as a promising solution for generating synthetic polyp images, but the image generation process in current models mainly relies on segmentation masks as the condition, limiting their ability to capture the full clinical context. To overcome these limitations, we propose a Progressive Spectrum Diffusion Model (PSDM) that integrates diverse clinical annotations-such as segmentation masks, bounding boxes, and colonoscopy reports-by transforming them into compositional prompts. These prompts are organized into coarse and fine components, allowing the model to capture both broad spatial structures and fine details, generating clinically accurate synthetic images. By augmenting training data with PSDM-generated samples, our model significantly improves polyp detection, classification, and segmentation. For instance, on the PolypGen dataset, PSDM increases the F1 score by 2.12% and the mean average precision by 3.09%, demonstrating superior performance in OOD scenarios and enhanced generalization.
Exploring Task-Level Optimal Prompts for Visual In-Context Learning
Jan 15, 2025Abstract:With the development of Vision Foundation Models (VFMs) in recent years, Visual In-Context Learning (VICL) has become a better choice compared to modifying models in most scenarios. Different from retraining or fine-tuning model, VICL does not require modifications to the model's weights or architecture, and only needs a prompt with demonstrations to teach VFM how to solve tasks. Currently, significant computational cost for finding optimal prompts for every test sample hinders the deployment of VICL, as determining which demonstrations to use for constructing prompts is very costly. In this paper, however, we find a counterintuitive phenomenon that most test samples actually achieve optimal performance under the same prompts, and searching for sample-level prompts only costs more time but results in completely identical prompts. Therefore, we propose task-level prompting to reduce the cost of searching for prompts during the inference stage and introduce two time-saving yet effective task-level prompt search strategies. Extensive experimental results show that our proposed method can identify near-optimal prompts and reach the best VICL performance with a minimal cost that prior work has never achieved.
Transformer-based toxin-protein interaction analysis prioritizes airborne particulate matter components with potential adverse health effects
Dec 21, 2024Abstract:Air pollution, particularly airborne particulate matter (PM), poses a significant threat to public health globally. It is crucial to comprehend the association between PM-associated toxic components and their cellular targets in humans to understand the mechanisms by which air pollution impacts health and to establish causal relationships between air pollution and public health consequences. Although many studies have explored the impact of PM on human health, the understanding of the association between toxins and the associated targets remain limited. Leveraging cutting-edge deep learning technologies, we developed tipFormer (toxin-protein interaction prediction based on transformer), a novel deep-learning tool for identifying toxic components capable of penetrating human cells and instigating pathogenic biological activities and signaling cascades. Experimental results show that tipFormer effectively captures interactions between proteins and toxic components. It incorporates dual pre-trained language models to encode protein sequences and chemicals. It employs a convolutional encoder to assimilate the sequential attributes of proteins and chemicals. It then introduces a learning module with a cross-attention mechanism to decode and elucidate the multifaceted interactions pivotal for the hotspots binding proteins and chemicals. Experimental results show that tipFormer effectively captures interactions between proteins and toxic components. This approach offers significant value to air quality and toxicology researchers by allowing high-throughput identification and prioritization of hazards. It supports more targeted laboratory studies and field measurements, ultimately enhancing our understanding of how air pollution impacts human health.
Exact: Exploring Space-Time Perceptive Clues for Weakly Supervised Satellite Image Time Series Semantic Segmentation
Dec 05, 2024



Abstract:Automated crop mapping through Satellite Image Time Series (SITS) has emerged as a crucial avenue for agricultural monitoring and management. However, due to the low resolution and unclear parcel boundaries, annotating pixel-level masks is exceptionally complex and time-consuming in SITS. This paper embraces the weakly supervised paradigm (i.e., only image-level categories available) to liberate the crop mapping task from the exhaustive annotation burden. The unique characteristics of SITS give rise to several challenges in weakly supervised learning: (1) noise perturbation from spatially neighboring regions, and (2) erroneous semantic bias from anomalous temporal periods. To address the above difficulties, we propose a novel method, termed exploring space-time perceptive clues (Exact). First, we introduce a set of spatial clues to explicitly capture the representative patterns of different crops from the most class-relative regions. Besides, we leverage the temporal-to-class interaction of the model to emphasize the contributions of pivotal clips, thereby enhancing the model perception for crop regions. Build upon the space-time perceptive clues, we derive the clue-based CAMs to effectively supervise the SITS segmentation network. Our method demonstrates impressive performance on various SITS benchmarks. Remarkably, the segmentation network trained on Exact-generated masks achieves 95% of its fully supervised performance, showing the bright promise of weakly supervised paradigm in crop mapping scenario. Our code will be publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge