Combining unsupervised and supervised learning for predicting the final stroke lesion
Jan 02, 2021Adriano Pinto, Sérgio Pereira, Raphael Meier, Roland Wiest, Victor Alves, Mauricio Reyes, Carlos A. Silva
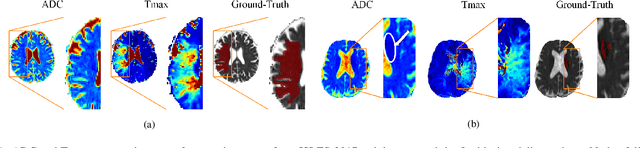

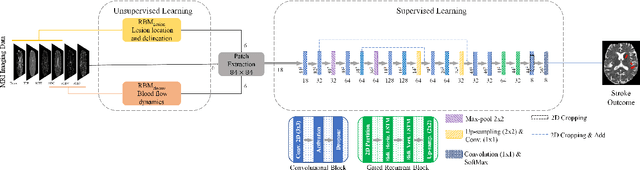
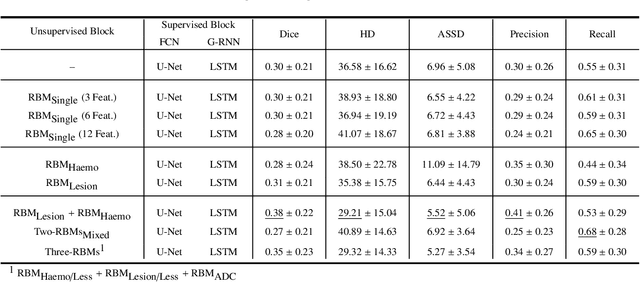
Predicting the final ischaemic stroke lesion provides crucial information regarding the volume of salvageable hypoperfused tissue, which helps physicians in the difficult decision-making process of treatment planning and intervention. Treatment selection is influenced by clinical diagnosis, which requires delineating the stroke lesion, as well as characterising cerebral blood flow dynamics using neuroimaging acquisitions. Nonetheless, predicting the final stroke lesion is an intricate task, due to the variability in lesion size, shape, location and the underlying cerebral haemodynamic processes that occur after the ischaemic stroke takes place. Moreover, since elapsed time between stroke and treatment is related to the loss of brain tissue, assessing and predicting the final stroke lesion needs to be performed in a short period of time, which makes the task even more complex. Therefore, there is a need for automatic methods that predict the final stroke lesion and support physicians in the treatment decision process. We propose a fully automatic deep learning method based on unsupervised and supervised learning to predict the final stroke lesion after 90 days. Our aim is to predict the final stroke lesion location and extent, taking into account the underlying cerebral blood flow dynamics that can influence the prediction. To achieve this, we propose a two-branch Restricted Boltzmann Machine, which provides specialized data-driven features from different sets of standard parametric Magnetic Resonance Imaging maps. These data-driven feature maps are then combined with the parametric Magnetic Resonance Imaging maps, and fed to a Convolutional and Recurrent Neural Network architecture. We evaluated our proposal on the publicly available ISLES 2017 testing dataset, reaching a Dice score of 0.38, Hausdorff Distance of 29.21 mm, and Average Symmetric Surface Distance of 5.52 mm.
Uncertainty-driven refinement of tumor-core segmentation using 3D-to-2D networks with label uncertainty
Dec 11, 2020Richard McKinley, Micheal Rebsamen, Katrin Daetwyler, Raphael Meier, Piotr Radojewski, Roland Wiest
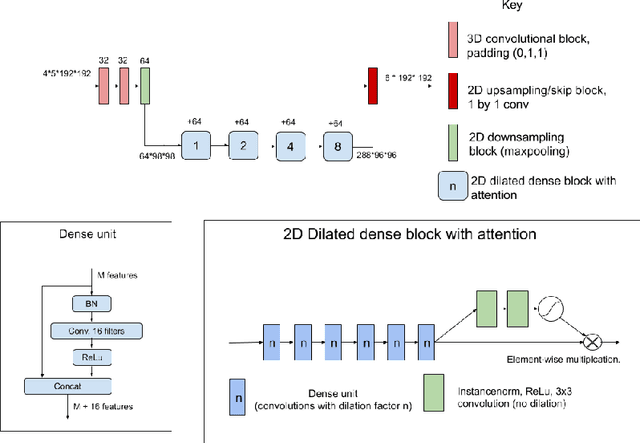
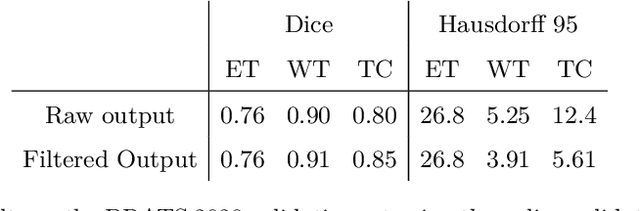
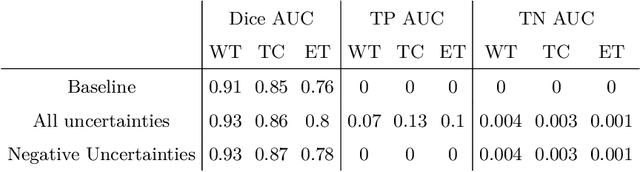
The BraTS dataset contains a mixture of high-grade and low-grade gliomas, which have a rather different appearance: previous studies have shown that performance can be improved by separated training on low-grade gliomas (LGGs) and high-grade gliomas (HGGs), but in practice this information is not available at test time to decide which model to use. By contrast with HGGs, LGGs often present no sharp boundary between the tumor core and the surrounding edema, but rather a gradual reduction of tumor-cell density. Utilizing our 3D-to-2D fully convolutional architecture, DeepSCAN, which ranked highly in the 2019 BraTS challenge and was trained using an uncertainty-aware loss, we separate cases into those with a confidently segmented core, and those with a vaguely segmented or missing core. Since by assumption every tumor has a core, we reduce the threshold for classification of core tissue in those cases where the core, as segmented by the classifier, is vaguely defined or missing. We then predict survival of high-grade glioma patients using a fusion of linear regression and random forest classification, based on age, number of distinct tumor components, and number of distinct tumor cores. We present results on the validation dataset of the Multimodal Brain Tumor Segmentation Challenge 2020 (segmentation and uncertainty challenge), and on the testing set, where the method achieved 4th place in Segmentation, 1st place in uncertainty estimation, and 1st place in Survival prediction.
Dual-Stream Pyramid Registration Network
Sep 26, 2019Xiaojun Hu, Miao Kang, Weilin Huang, Matthew R. Scott, Roland Wiest, Mauricio Reyes
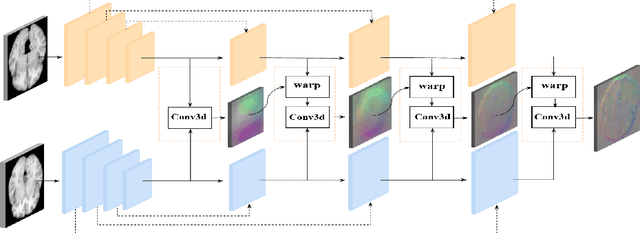
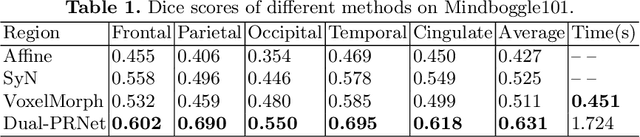
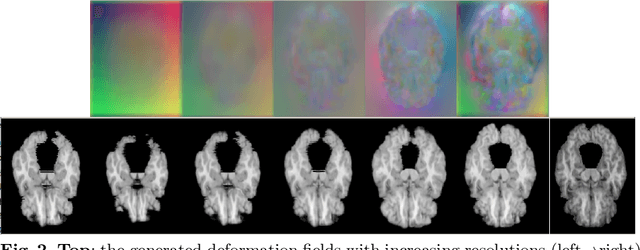
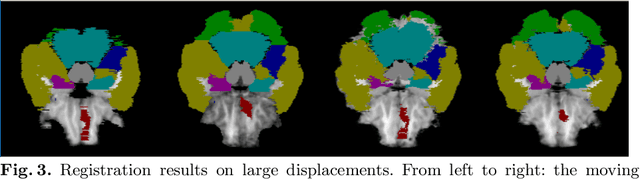
We propose a Dual-Stream Pyramid Registration Network (referred as Dual-PRNet) for unsupervised 3D medical image registration. Unlike recent CNN-based registration approaches, such as VoxelMorph, which explores a single-stream encoder-decoder network to compute a registration fields from a pair of 3D volumes, we design a two-stream architecture able to compute multi-scale registration fields from convolutional feature pyramids. Our contributions are two-fold: (i) we design a two-stream 3D encoder-decoder network which computes two convolutional feature pyramids separately for a pair of input volumes, resulting in strong deep representations that are meaningful for deformation estimation; (ii) we propose a pyramid registration module able to predict multi-scale registration fields directly from the decoding feature pyramids. This allows it to refine the registration fields gradually in a coarse-to-fine manner via sequential warping, and enable the model with the capability for handling significant deformations between two volumes, such as large displacements in spatial domain or slice space. The proposed Dual-PRNet is evaluated on two standard benchmarks for brain MRI registration, where it outperforms the state-of-the-art approaches by a large margin, e.g., having improvements over recent VoxelMorph [2] with 0.683->0.778 on the LPBA40, and 0.511->0.631 on the Mindboggle101, in term of average Dice score.
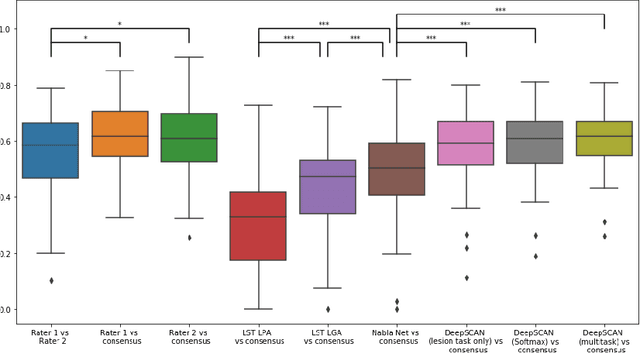
Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence
Apr 05, 2019Richard McKinley, Lorenz Grunder, Rik Wepfer, Fabian Aschwanden, Tim Fischer, Christoph Friedli, Raphaela Muri, Christian Rummel, Rajeev Verma, Christian Weisstanner, Mauricio Reyes, Anke Salmen, Andrew Chan, Roland Wiest, Franca Wagner
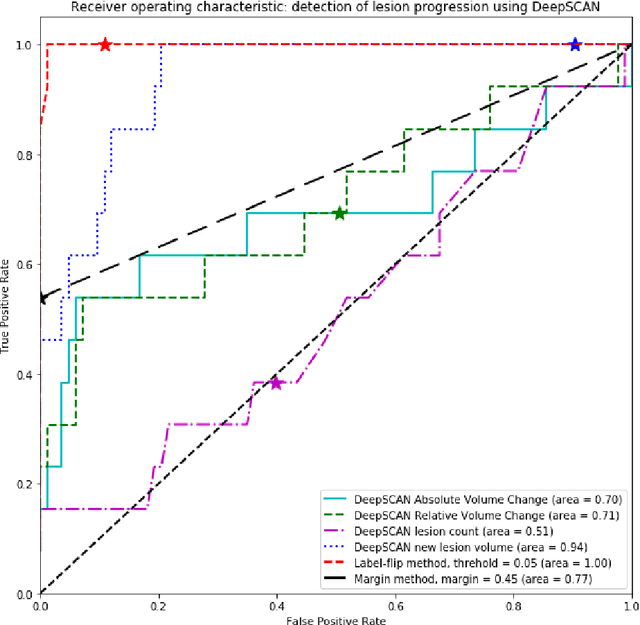
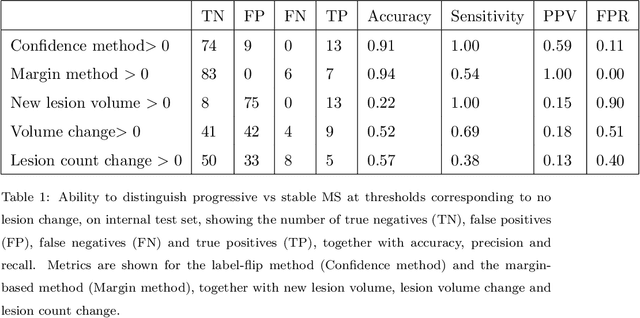
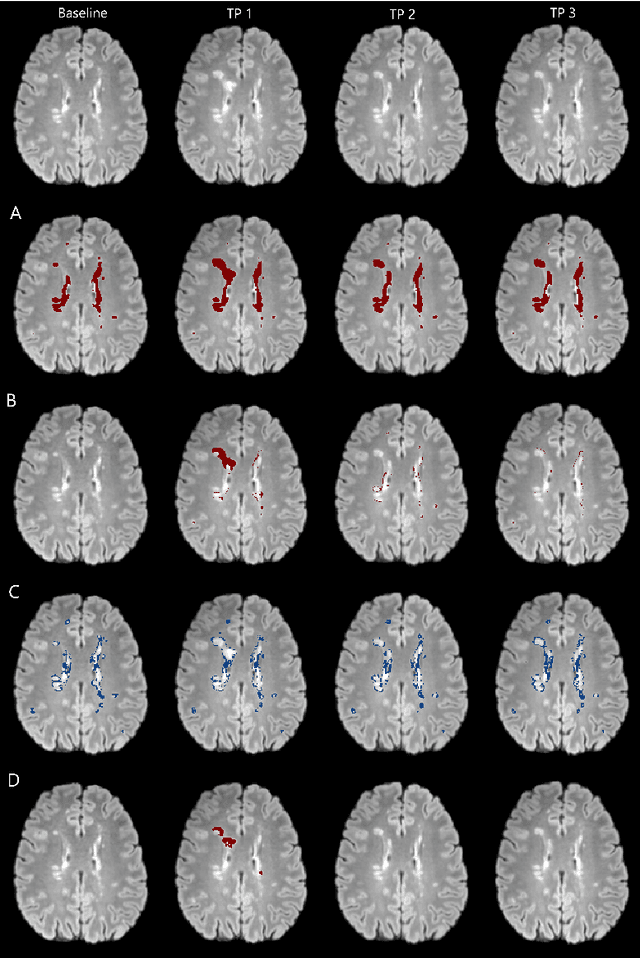
The detection of new or enlarged white-matter lesions in multiple sclerosis is a vital task in the monitoring of patients undergoing disease-modifying treatment for multiple sclerosis. However, the definition of 'new or enlarged' is not fixed, and it is known that lesion-counting is highly subjective, with high degree of inter- and intra-rater variability. Automated methods for lesion quantification hold the potential to make the detection of new and enlarged lesions consistent and repeatable. However, the majority of lesion segmentation algorithms are not evaluated for their ability to separate progressive from stable patients, despite this being a pressing clinical use-case. In this paper we show that change in volumetric measurements of lesion load alone is not a good method for performing this separation, even for highly performing segmentation methods. Instead, we propose a method for identifying lesion changes of high certainty, and establish on a dataset of longitudinal multiple sclerosis cases that this method is able to separate progressive from stable timepoints with a very high level of discrimination (AUC = 0.99), while changes in lesion volume are much less able to perform this separation (AUC = 0.71). Validation of the method on a second external dataset confirms that the method is able to generalize beyond the setting in which it was trained, achieving an accuracy of 83% in separating stable and progressive timepoints. Both lesion volume and count have previously been shown to be strong predictors of disease course across a population. However, we demonstrate that for individual patients, changes in these measures are not an adequate means of establishing no evidence of disease activity. Meanwhile, directly detecting tissue which changes, with high confidence, from non-lesion to lesion is a feasible methodology for identifying radiologically active patients.
Few-shot brain segmentation from weakly labeled data with deep heteroscedastic multi-task networks
Apr 04, 2019Richard McKinley, Michael Rebsamen, Raphael Meier, Mauricio Reyes, Christian Rummel, Roland Wiest
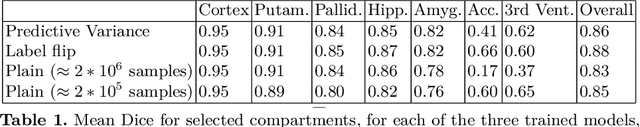
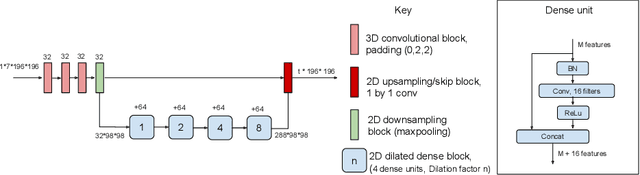
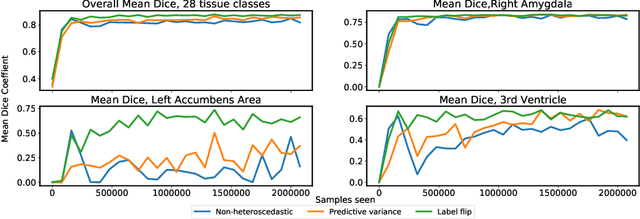
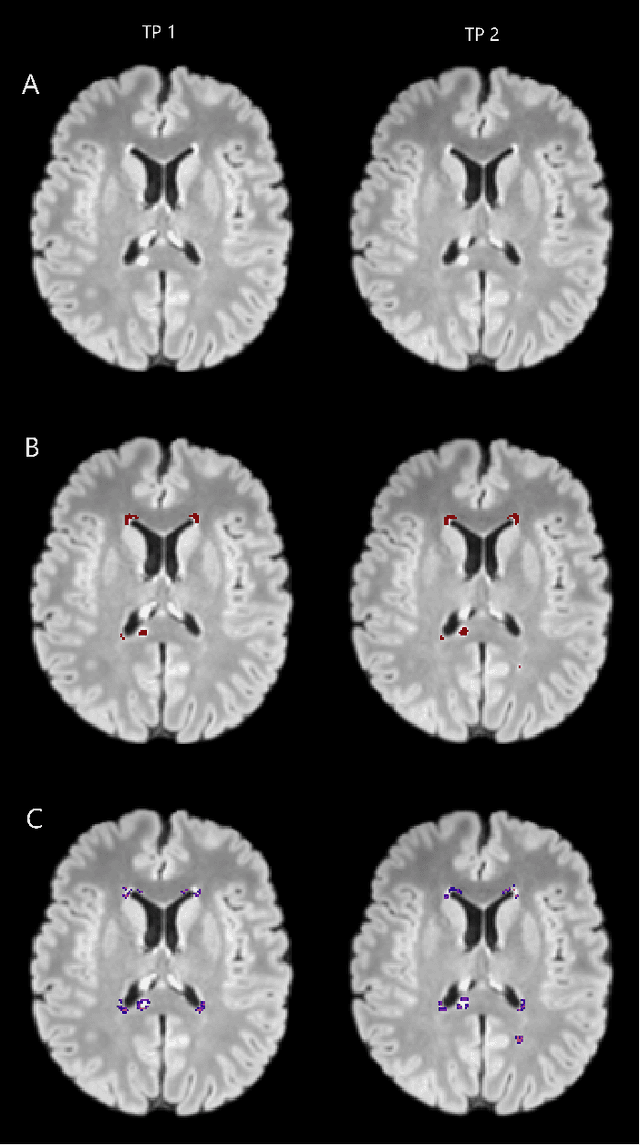
In applications of supervised learning applied to medical image segmentation, the need for large amounts of labeled data typically goes unquestioned. In particular, in the case of brain anatomy segmentation, hundreds or thousands of weakly-labeled volumes are often used as training data. In this paper, we first observe that for many brain structures, a small number of training examples, (n=9), weakly labeled using Freesurfer 6.0, plus simple data augmentation, suffice as training data to achieve high performance, achieving an overall mean Dice coefficient of $0.84 \pm 0.12$ compared to Freesurfer over 28 brain structures in T1-weighted images of $\approx 4000$ 9-10 year-olds from the Adolescent Brain Cognitive Development study. We then examine two varieties of heteroscedastic network as a method for improving classification results. An existing proposal by Kendall and Gal, which uses Monte-Carlo inference to learn to predict the variance of each prediction, yields an overall mean Dice of $0.85 \pm 0.14$ and showed statistically significant improvements over 25 brain structures. Meanwhile a novel heteroscedastic network which directly learns the probability that an example has been mislabeled yielded an overall mean Dice of $0.87 \pm 0.11$ and showed statistically significant improvements over all but one of the brain structures considered. The loss function associated to this network can be interpreted as performing a form of learned label smoothing, where labels are only smoothed where they are judged to be uncertain.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019Hugo J. Kuijf, J. Matthijs Biesbroek, Jeroen de Bresser, Rutger Heinen, Simon Andermatt, Mariana Bento, Matt Berseth, Mikhail Belyaev, M. Jorge Cardoso, Adrià Casamitjana, D. Louis Collins, Mahsa Dadar, Achilleas Georgiou, Mohsen Ghafoorian, Dakai Jin, April Khademi, Jesse Knight, Hongwei Li, Xavier Lladó, Miguel Luna, Qaiser Mahmood, Richard McKinley, Alireza Mehrtash, Sébastien Ourselin, Bo-yong Park, Hyunjin Park, Sang Hyun Park, Simon Pezold, Elodie Puybareau, Leticia Rittner, Carole H. Sudre, Sergi Valverde, Verónica Vilaplana, Roland Wiest, Yongchao Xu, Ziyue Xu, Guodong Zeng, Jianguo Zhang, Guoyan Zheng, Christopher Chen, Wiesje van der Flier, Frederik Barkhof, Max A. Viergever, Geert Jan Biessels
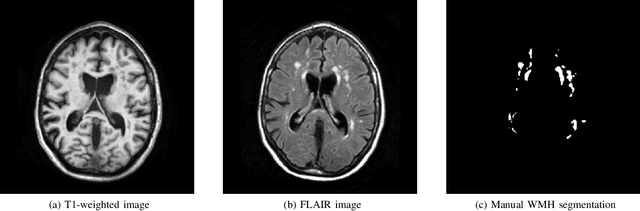
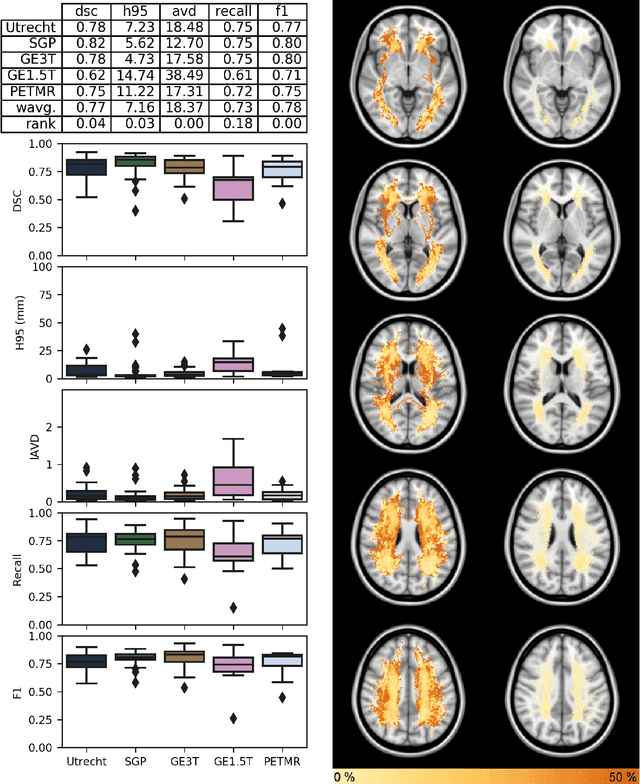
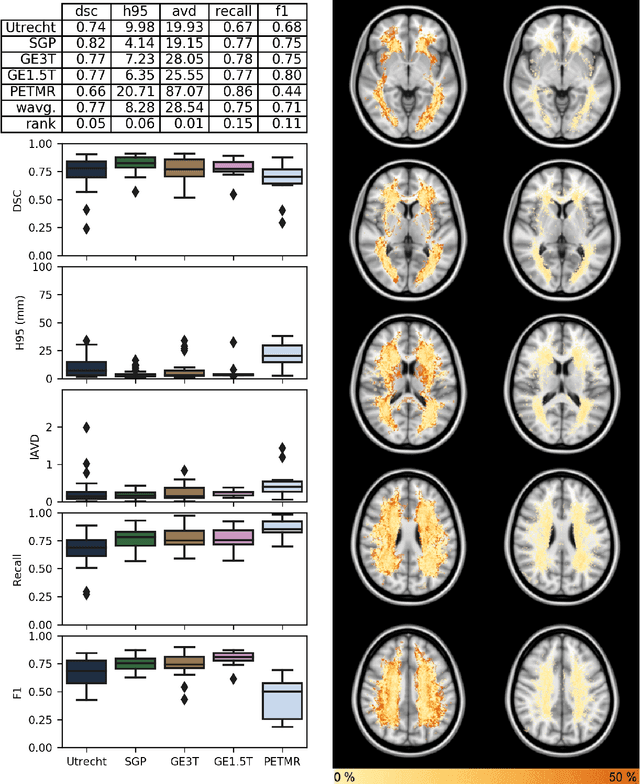
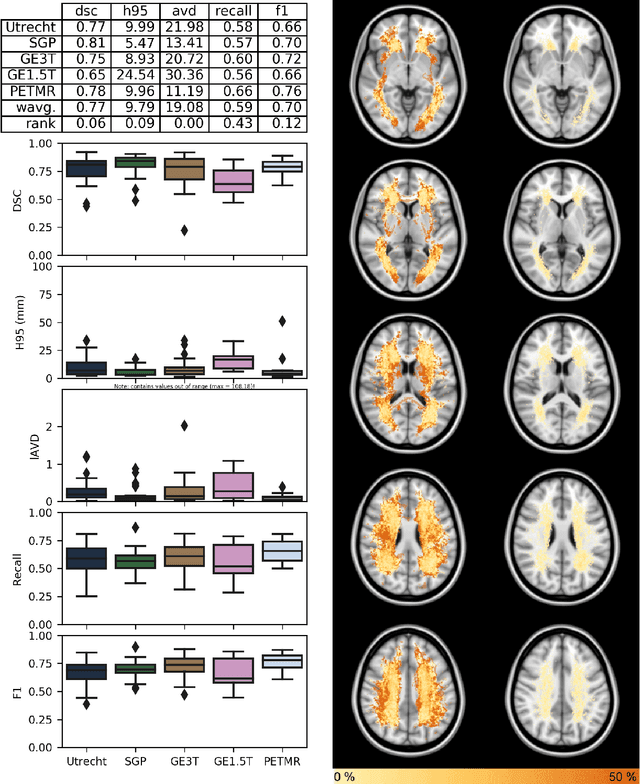
Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Simultaneous lesion and neuroanatomy segmentation in Multiple Sclerosis using deep neural networks
Jan 22, 2019Richard McKinley, Rik Wepfer, Fabian Aschwanden, Lorenz Grunder, Raphaela Muri, Christian Rummel, Rajeev Verma, Christian Weisstanner, Mauricio Reyes, Anke Salmen, Andrew Chan, Franca Wagner, Roland Wiest
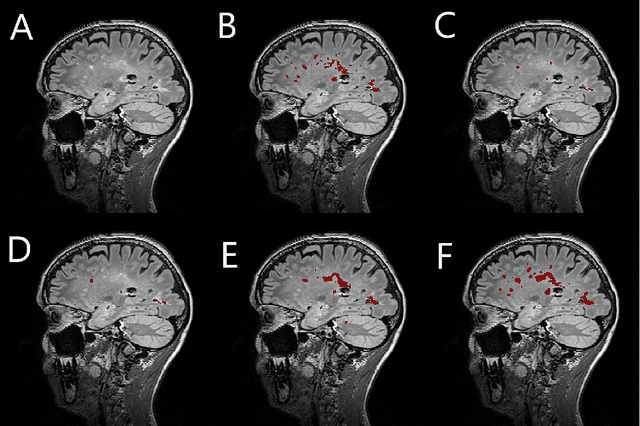
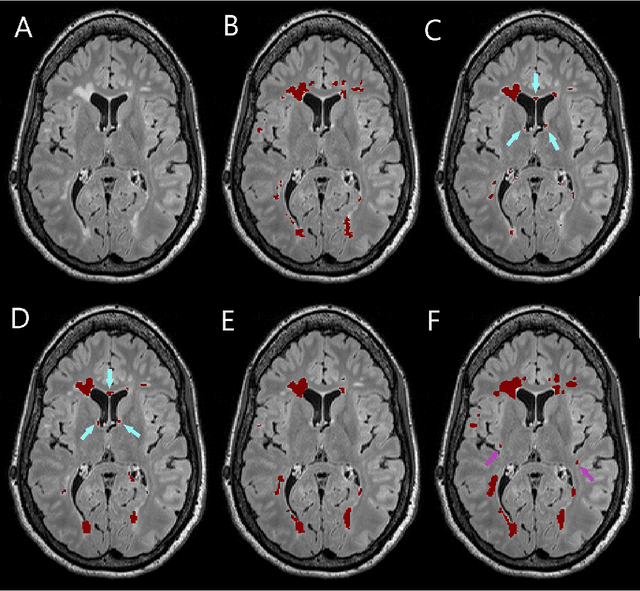
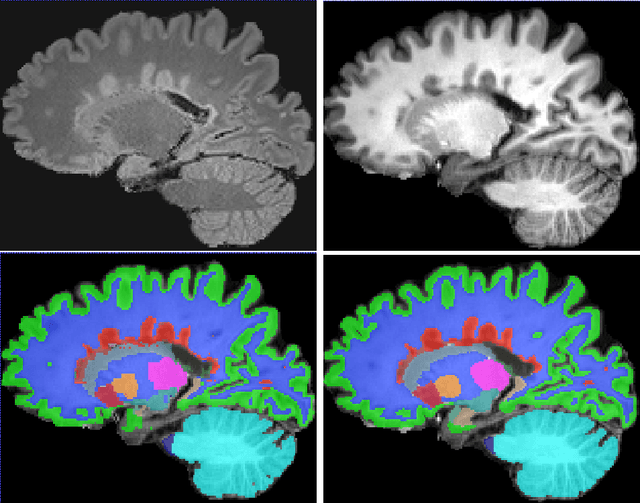
Segmentation of both white matter lesions and deep grey matter structures is an important task in the quantification of magnetic resonance imaging in multiple sclerosis. Typically these tasks are performed separately: in this paper we present a single CNN-based segmentation solution for providing fast, reliable segmentations of multimodal MR imagies into lesion classes and healthy-appearing grey- and white-matter structures. We show substantial, statistically significant improvements in both Dice coefficient and in lesion-wise specificity and sensitivity, compared to previous approaches, and agreement with individual human raters in the range of human inter-rater variability. The method is trained on data gathered from a single centre: nonetheless, it performs well on data from centres, scanners and field-strengths not represented in the training dataset. A retrospective study found that the classifier successfully identified lesions missed by the human raters. Lesion labels were provided by human raters, while weak labels for other brain structures (including CSF, cortical grey matter, cortical white matter, cerebellum, amygdala, hippocampus, subcortical GM structures and choroid plexus) were provided by Freesurfer 5.3. The segmentations of these structures compared well, not only with Freesurfer 5.3, but also with FSL-First and Freesurfer 6.1.
Deep Learning versus Classical Regression for Brain Tumor Patient Survival Prediction
Nov 12, 2018Yannick Suter, Alain Jungo, Michael Rebsamen, Urspeter Knecht, Evelyn Herrmann, Roland Wiest, Mauricio Reyes
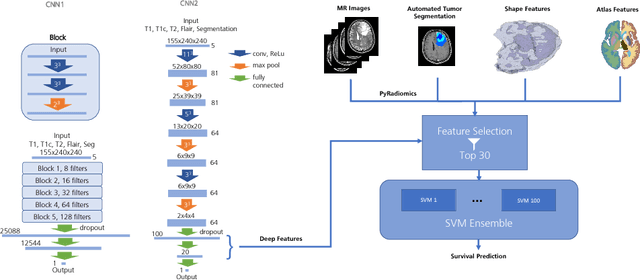
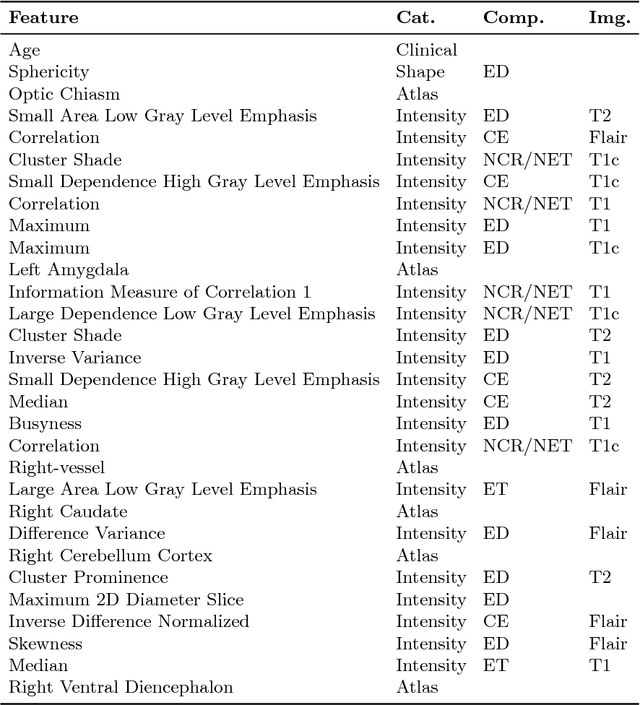
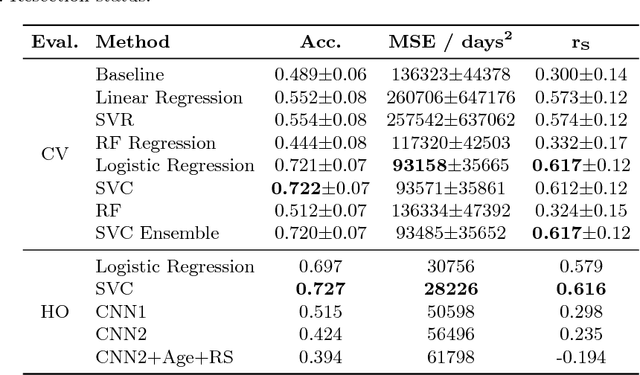
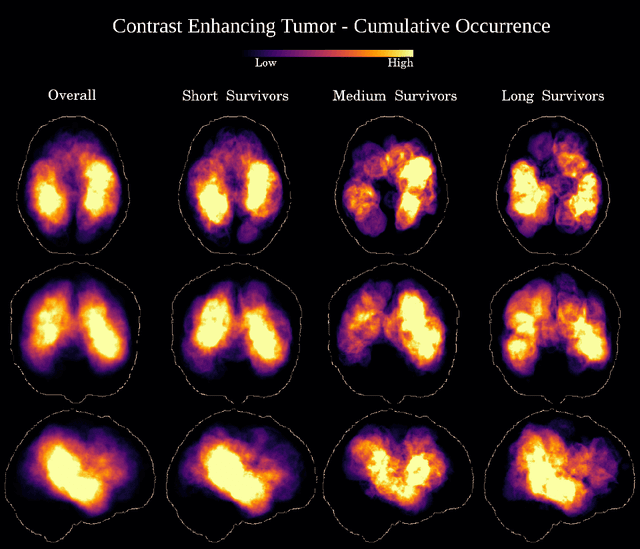
Deep learning for regression tasks on medical imaging data has shown promising results. However, compared to other approaches, their power is strongly linked to the dataset size. In this study, we evaluate 3D-convolutional neural networks (CNNs) and classical regression methods with hand-crafted features for survival time regression of patients with high grade brain tumors. The tested CNNs for regression showed promising but unstable results. The best performing deep learning approach reached an accuracy of 51.5% on held-out samples of the training set. All tested deep learning experiments were outperformed by a Support Vector Classifier (SVC) using 30 radiomic features. The investigated features included intensity, shape, location and deep features. The submitted method to the BraTS 2018 survival prediction challenge is an ensemble of SVCs, which reached a cross-validated accuracy of 72.2% on the BraTS 2018 training set, 57.1% on the validation set, and 42.9% on the testing set. The results suggest that more training data is necessary for a stable performance of a CNN model for direct regression from magnetic resonance images, and that non-imaging clinical patient information is crucial along with imaging information.
Enhancing clinical MRI Perfusion maps with data-driven maps of complementary nature for lesion outcome prediction
Jun 12, 2018Adriano Pinto, Sergio Pereira, Raphael Meier, Victor Alves, Roland Wiest, Carlos A. Silva, Mauricio Reyes
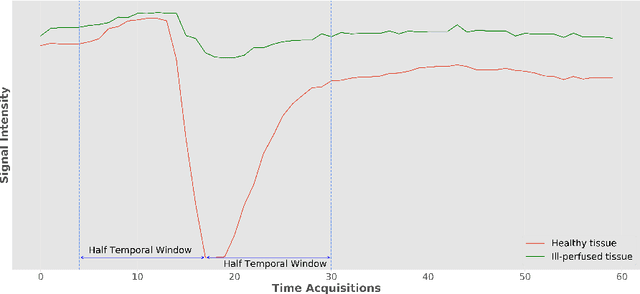
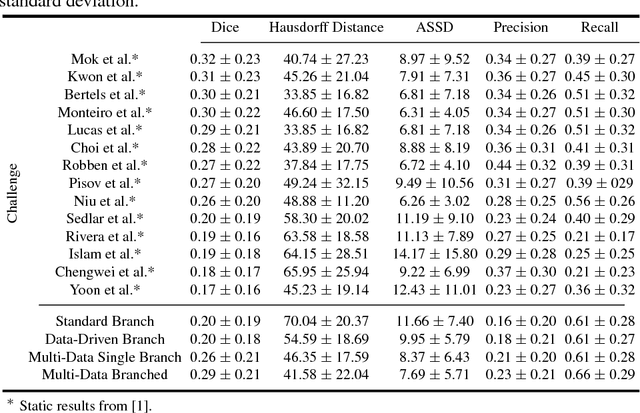
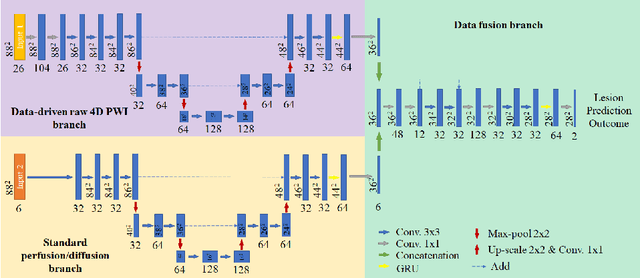
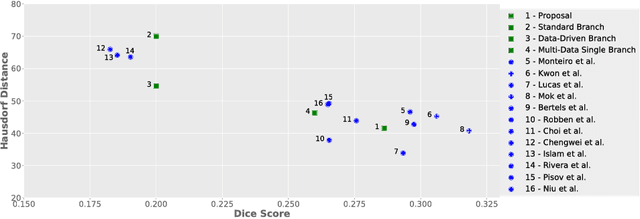
Stroke is the second most common cause of death in developed countries, where rapid clinical intervention can have a major impact on a patient's life. To perform the revascularization procedure, the decision making of physicians considers its risks and benefits based on multi-modal MRI and clinical experience. Therefore, automatic prediction of the ischemic stroke lesion outcome has the potential to assist the physician towards a better stroke assessment and information about tissue outcome. Typically, automatic methods consider the information of the standard kinetic models of diffusion and perfusion MRI (e.g. Tmax, TTP, MTT, rCBF, rCBV) to perform lesion outcome prediction. In this work, we propose a deep learning method to fuse this information with an automated data selection of the raw 4D PWI image information, followed by a data-driven deep-learning modeling of the underlying blood flow hemodynamics. We demonstrate the ability of the proposed approach to improve prediction of tissue at risk before therapy, as compared to only using the standard clinical perfusion maps, hence suggesting on the potential benefits of the proposed data-driven raw perfusion data modelling approach.
Synthetic Perfusion Maps: Imaging Perfusion Deficits in DSC-MRI with Deep Learning
Jun 11, 2018Andreas Hess, Raphael Meier, Johannes Kaesmacher, Simon Jung, Fabien Scalzo, David Liebeskind, Roland Wiest, Richard McKinley
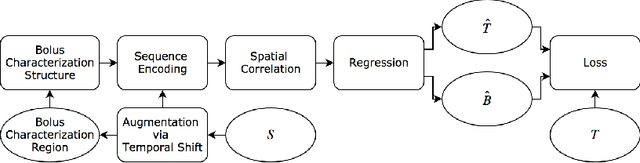
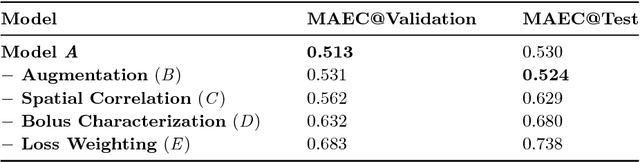
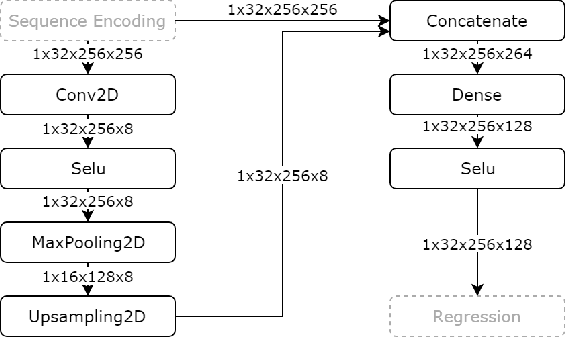
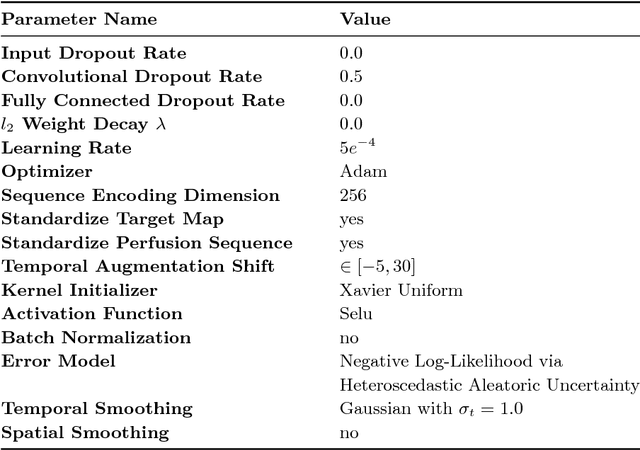
In this work, we present a novel convolutional neural net- work based method for perfusion map generation in dynamic suscepti- bility contrast-enhanced perfusion imaging. The proposed architecture is trained end-to-end and solely relies on raw perfusion data for inference. We used a dataset of 151 acute ischemic stroke cases for evaluation. Our method generates perfusion maps that are comparable to the target maps used for clinical routine, while being model-free, fast, and less noisy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge