Cuntai Guan
Decoupled Hierarchical Distillation for Multimodal Emotion Recognition
Feb 04, 2026Abstract:Human multimodal emotion recognition (MER) seeks to infer human emotions by integrating information from language, visual, and acoustic modalities. Although existing MER approaches have achieved promising results, they still struggle with inherent multimodal heterogeneities and varying contributions from different modalities. To address these challenges, we propose a novel framework, Decoupled Hierarchical Multimodal Distillation (DHMD). DHMD decouples each modality's features into modality-irrelevant (homogeneous) and modality-exclusive (heterogeneous) components using a self-regression mechanism. The framework employs a two-stage knowledge distillation (KD) strategy: (1) coarse-grained KD via a Graph Distillation Unit (GD-Unit) in each decoupled feature space, where a dynamic graph facilitates adaptive distillation among modalities, and (2) fine-grained KD through a cross-modal dictionary matching mechanism, which aligns semantic granularities across modalities to produce more discriminative MER representations. This hierarchical distillation approach enables flexible knowledge transfer and effectively improves cross-modal feature alignment. Experimental results demonstrate that DHMD consistently outperforms state-of-the-art MER methods, achieving 1.3\%/2.4\% (ACC$_7$), 1.3\%/1.9\% (ACC$_2$) and 1.9\%/1.8\% (F1) relative improvement on CMU-MOSI/CMU-MOSEI dataset, respectively. Meanwhile, visualization results reveal that both the graph edges and dictionary activations in DHMD exhibit meaningful distribution patterns across modality-irrelevant/-exclusive feature spaces.
* arXiv admin note: text overlap with arXiv:2303.13802
DepFlow: Disentangled Speech Generation to Mitigate Semantic Bias in Depression Detection
Jan 01, 2026Abstract:Speech is a scalable and non-invasive biomarker for early mental health screening. However, widely used depression datasets like DAIC-WOZ exhibit strong coupling between linguistic sentiment and diagnostic labels, encouraging models to learn semantic shortcuts. As a result, model robustness may be compromised in real-world scenarios, such as Camouflaged Depression, where individuals maintain socially positive or neutral language despite underlying depressive states. To mitigate this semantic bias, we propose DepFlow, a three-stage depression-conditioned text-to-speech framework. First, a Depression Acoustic Encoder learns speaker- and content-invariant depression embeddings through adversarial training, achieving effective disentanglement while preserving depression discriminability (ROC-AUC: 0.693). Second, a flow-matching TTS model with FiLM modulation injects these embeddings into synthesis, enabling control over depressive severity while preserving content and speaker identity. Third, a prototype-based severity mapping mechanism provides smooth and interpretable manipulation across the depression continuum. Using DepFlow, we construct a Camouflage Depression-oriented Augmentation (CDoA) dataset that pairs depressed acoustic patterns with positive/neutral content from a sentiment-stratified text bank, creating acoustic-semantic mismatches underrepresented in natural data. Evaluated across three depression detection architectures, CDoA improves macro-F1 by 9%, 12%, and 5%, respectively, consistently outperforming conventional augmentation strategies in depression Detection. Beyond enhancing robustness, DepFlow provides a controllable synthesis platform for conversational systems and simulation-based evaluation, where real clinical data remains limited by ethical and coverage constraints.
EEG-DLite: Dataset Distillation for Efficient Large EEG Model Training
Dec 13, 2025



Abstract:Large-scale EEG foundation models have shown strong generalization across a range of downstream tasks, but their training remains resource-intensive due to the volume and variable quality of EEG data. In this work, we introduce EEG-DLite, a data distillation framework that enables more efficient pre-training by selectively removing noisy and redundant samples from large EEG datasets. EEG-DLite begins by encoding EEG segments into compact latent representations using a self-supervised autoencoder, allowing sample selection to be performed efficiently and with reduced sensitivity to noise. Based on these representations, EEG-DLite filters out outliers and minimizes redundancy, resulting in a smaller yet informative subset that retains the diversity essential for effective foundation model training. Through extensive experiments, we demonstrate that training on only 5 percent of a 2,500-hour dataset curated with EEG-DLite yields performance comparable to, and in some cases better than, training on the full dataset across multiple downstream tasks. To our knowledge, this is the first systematic study of pre-training data distillation in the context of EEG foundation models. EEG-DLite provides a scalable and practical path toward more effective and efficient physiological foundation modeling. The code is available at https://github.com/t170815518/EEG-DLite.
MATAI: A Generalist Machine Learning Framework for Property Prediction and Inverse Design of Advanced Alloys
Nov 13, 2025



Abstract:The discovery of advanced metallic alloys is hindered by vast composition spaces, competing property objectives, and real-world constraints on manufacturability. Here we introduce MATAI, a generalist machine learning framework for property prediction and inverse design of as-cast alloys. MATAI integrates a curated alloy database, deep neural network-based property predictors, a constraint-aware optimization engine, and an iterative AI-experiment feedback loop. The framework estimates key mechanical propertie, sincluding density, yield strength, ultimate tensile strength, and elongation, directly from composition, using multi-task learning and physics-informed inductive biases. Alloy design is framed as a constrained optimization problem and solved using a bi-level approach that combines local search with symbolic constraint programming. We demonstrate MATAI's capabilities on the Ti-based alloy system, a canonical class of lightweight structural materials, where it rapidly identifies candidates that simultaneously achieve lower density (<4.45 g/cm3), higher strength (>1000 MPa) and appreciable ductility (>5%) through only seven iterations. Experimental validation confirms that MATAI-designed alloys outperform commercial references such as TC4, highlighting the framework's potential to accelerate the discovery of lightweight, high-performance materials under real-world design constraints.
Unveiling Deep Semantic Uncertainty Perception for Language-Anchored Multi-modal Vision-Brain Alignment
Nov 06, 2025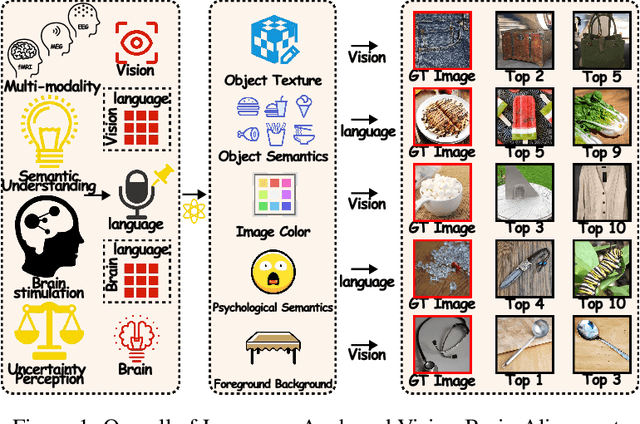
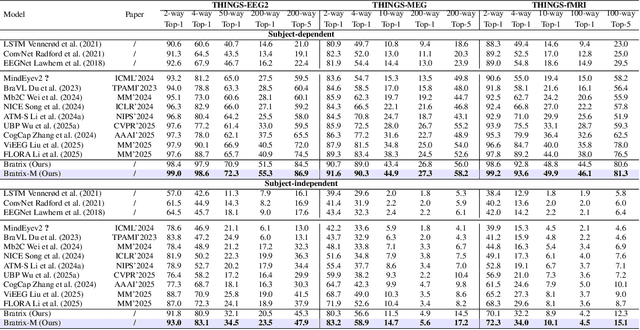
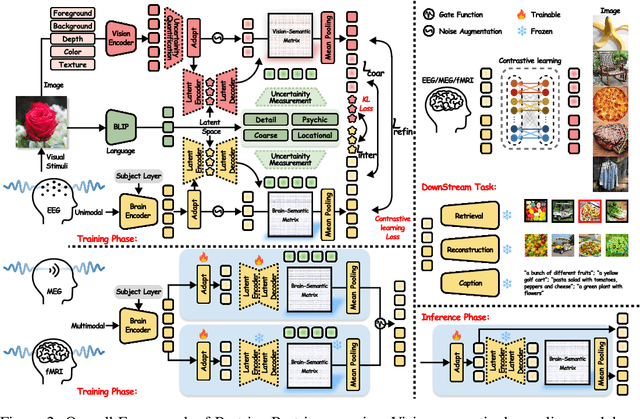
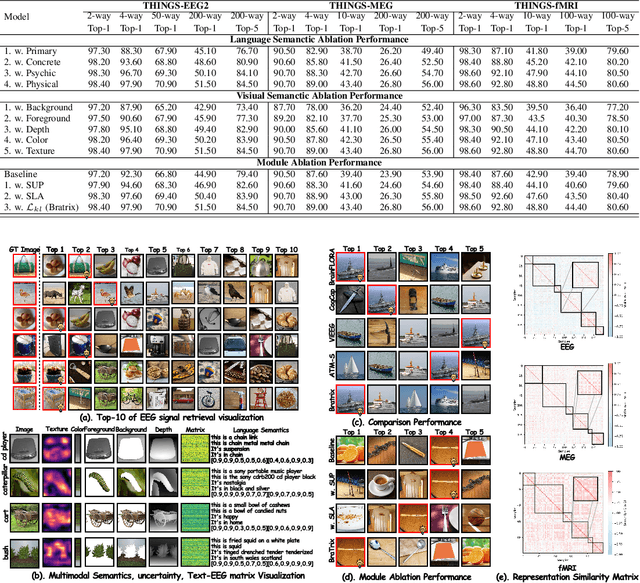
Abstract:Unveiling visual semantics from neural signals such as EEG, MEG, and fMRI remains a fundamental challenge due to subject variability and the entangled nature of visual features. Existing approaches primarily align neural activity directly with visual embeddings, but visual-only representations often fail to capture latent semantic dimensions, limiting interpretability and deep robustness. To address these limitations, we propose Bratrix, the first end-to-end framework to achieve multimodal Language-Anchored Vision-Brain alignment. Bratrix decouples visual stimuli into hierarchical visual and linguistic semantic components, and projects both visual and brain representations into a shared latent space, enabling the formation of aligned visual-language and brain-language embeddings. To emulate human-like perceptual reliability and handle noisy neural signals, Bratrix incorporates a novel uncertainty perception module that applies uncertainty-aware weighting during alignment. By leveraging learnable language-anchored semantic matrices to enhance cross-modal correlations and employing a two-stage training strategy of single-modality pretraining followed by multimodal fine-tuning, Bratrix-M improves alignment precision. Extensive experiments on EEG, MEG, and fMRI benchmarks demonstrate that Bratrix improves retrieval, reconstruction, and captioning performance compared to state-of-the-art methods, specifically surpassing 14.3% in 200-way EEG retrieval task. Code and model are available.
BrainPro: Towards Large-scale Brain State-aware EEG Representation Learning
Sep 26, 2025



Abstract:Electroencephalography (EEG) is a non-invasive technique for recording brain electrical activity, widely used in brain-computer interface (BCI) and healthcare. Recent EEG foundation models trained on large-scale datasets have shown improved performance and generalizability over traditional decoding methods, yet significant challenges remain. Existing models often fail to explicitly capture channel-to-channel and region-to-region interactions, which are critical sources of information inherently encoded in EEG signals. Due to varying channel configurations across datasets, they either approximate spatial structure with self-attention or restrict training to a limited set of common channels, sacrificing flexibility and effectiveness. Moreover, although EEG datasets reflect diverse brain states such as emotion, motor, and others, current models rarely learn state-aware representations during self-supervised pre-training. To address these gaps, we propose BrainPro, a large EEG model that introduces a retrieval-based spatial learning block to flexibly capture channel- and region-level interactions across varying electrode layouts, and a brain state-decoupling block that enables state-aware representation learning through parallel encoders with decoupling and region-aware reconstruction losses. This design allows BrainPro to adapt seamlessly to diverse tasks and hardware settings. Pre-trained on an extensive EEG corpus, BrainPro achieves state-of-the-art performance and robust generalization across nine public BCI datasets. Our codes and the pre-trained weights will be released.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Towards Robust Multimodal Physiological Foundation Models: Handling Arbitrary Missing Modalities
Apr 28, 2025



Abstract:Multimodal physiological signals, such as EEG, ECG, EOG, and EMG, are crucial for healthcare and brain-computer interfaces. While existing methods rely on specialized architectures and dataset-specific fusion strategies, they struggle to learn universal representations that generalize across datasets and handle missing modalities at inference time. To address these issues, we propose PhysioOmni, a foundation model for multimodal physiological signal analysis that models both homogeneous and heterogeneous features to decouple multimodal signals and extract generic representations while maintaining compatibility with arbitrary missing modalities. PhysioOmni trains a decoupled multimodal tokenizer, enabling masked signal pre-training via modality-invariant and modality-specific objectives. To ensure adaptability to diverse and incomplete modality combinations, the pre-trained encoders undergo resilient fine-tuning with prototype alignment on downstream datasets. Extensive experiments on four downstream tasks, emotion recognition, sleep stage classification, motor prediction, and mental workload detection, demonstrate that PhysioOmni achieves state-of-the-art performance while maintaining strong robustness to missing modalities. Our code and model weights will be released.
SPD Learning for Covariance-Based Neuroimaging Analysis: Perspectives, Methods, and Challenges
Apr 26, 2025



Abstract:Neuroimaging provides a critical framework for characterizing brain activity by quantifying connectivity patterns and functional architecture across modalities. While modern machine learning has significantly advanced our understanding of neural processing mechanisms through these datasets, decoding task-specific signatures must contend with inherent neuroimaging constraints, for example, low signal-to-noise ratios in raw electrophysiological recordings, cross-session non-stationarity, and limited sample sizes. This review focuses on machine learning approaches for covariance-based neuroimaging data, where often symmetric positive definite (SPD) matrices under full-rank conditions encode inter-channel relationships. By equipping the space of SPD matrices with Riemannian metrics (e.g., affine-invariant or log-Euclidean), their space forms a Riemannian manifold enabling geometric analysis. We unify methodologies operating on this manifold under the SPD learning framework, which systematically leverages the SPD manifold's geometry to process covariance features, thereby advancing brain imaging analytics.
Beyond Overfitting: Doubly Adaptive Dropout for Generalizable AU Detection
Mar 12, 2025



Abstract:Facial Action Units (AUs) are essential for conveying psychological states and emotional expressions. While automatic AU detection systems leveraging deep learning have progressed, they often overfit to specific datasets and individual features, limiting their cross-domain applicability. To overcome these limitations, we propose a doubly adaptive dropout approach for cross-domain AU detection, which enhances the robustness of convolutional feature maps and spatial tokens against domain shifts. This approach includes a Channel Drop Unit (CD-Unit) and a Token Drop Unit (TD-Unit), which work together to reduce domain-specific noise at both the channel and token levels. The CD-Unit preserves domain-agnostic local patterns in feature maps, while the TD-Unit helps the model identify AU relationships generalizable across domains. An auxiliary domain classifier, integrated at each layer, guides the selective omission of domain-sensitive features. To prevent excessive feature dropout, a progressive training strategy is used, allowing for selective exclusion of sensitive features at any model layer. Our method consistently outperforms existing techniques in cross-domain AU detection, as demonstrated by extensive experimental evaluations. Visualizations of attention maps also highlight clear and meaningful patterns related to both individual and combined AUs, further validating the approach's effectiveness.
* Accetped by IEEE Transactions on Affective Computing 2025. A novel method for cross-domain facial action unit detection
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge