Hadrien Reynaud
InfoMotion: A Graph-Based Approach to Video Dataset Distillation for Echocardiography
Dec 13, 2025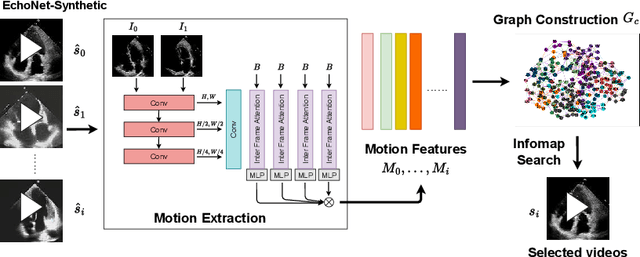
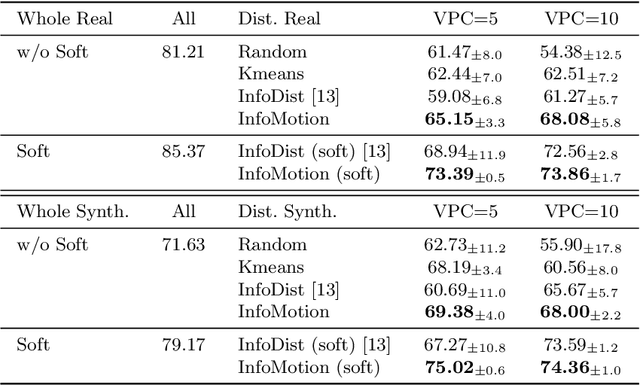
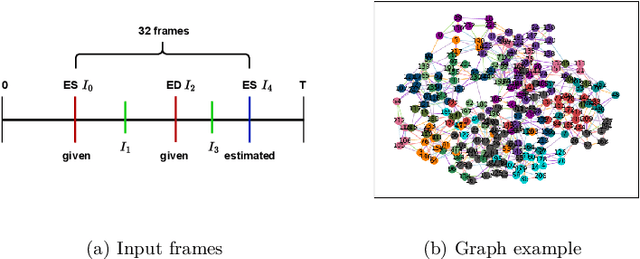
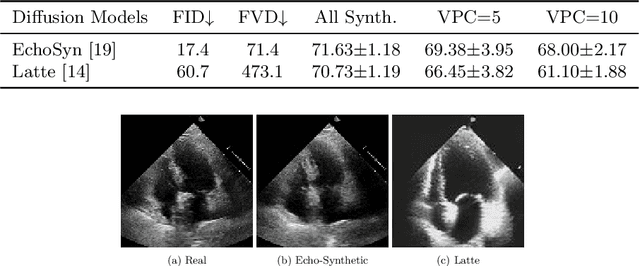
Abstract:Echocardiography plays a critical role in the diagnosis and monitoring of cardiovascular diseases as a non-invasive real-time assessment of cardiac structure and function. However, the growing scale of echocardiographic video data presents significant challenges in terms of storage, computation, and model training efficiency. Dataset distillation offers a promising solution by synthesizing a compact, informative subset of data that retains the key clinical features of the original dataset. In this work, we propose a novel approach for distilling a compact synthetic echocardiographic video dataset. Our method leverages motion feature extraction to capture temporal dynamics, followed by class-wise graph construction and representative sample selection using the Infomap algorithm. This enables us to select a diverse and informative subset of synthetic videos that preserves the essential characteristics of the original dataset. We evaluate our approach on the EchoNet-Dynamic datasets and achieve a test accuracy of \(69.38\%\) using only \(25\) synthetic videos. These results demonstrate the effectiveness and scalability of our method for medical video dataset distillation.
Label-free Motion-Conditioned Diffusion Model for Cardiac Ultrasound Synthesis
Dec 10, 2025Abstract:Ultrasound echocardiography is essential for the non-invasive, real-time assessment of cardiac function, but the scarcity of labelled data, driven by privacy restrictions and the complexity of expert annotation, remains a major obstacle for deep learning methods. We propose the Motion Conditioned Diffusion Model (MCDM), a label-free latent diffusion framework that synthesises realistic echocardiography videos conditioned on self-supervised motion features. To extract these features, we design the Motion and Appearance Feature Extractor (MAFE), which disentangles motion and appearance representations from videos. Feature learning is further enhanced by two auxiliary objectives: a re-identification loss guided by pseudo appearance features and an optical flow loss guided by pseudo flow fields. Evaluated on the EchoNet-Dynamic dataset, MCDM achieves competitive video generation performance, producing temporally coherent and clinically realistic sequences without reliance on manual labels. These results demonstrate the potential of self-supervised conditioning for scalable echocardiography synthesis. Our code is available at https://github.com/ZheLi2020/LabelfreeMCDM.
Better Tokens for Better 3D: Advancing Vision-Language Modeling in 3D Medical Imaging
Oct 23, 2025Abstract:Recent progress in vision-language modeling for 3D medical imaging has been fueled by large-scale computed tomography (CT) corpora with paired free-text reports, stronger architectures, and powerful pretrained models. This has enabled applications such as automated report generation and text-conditioned 3D image synthesis. Yet, current approaches struggle with high-resolution, long-sequence volumes: contrastive pretraining often yields vision encoders that are misaligned with clinical language, and slice-wise tokenization blurs fine anatomy, reducing diagnostic performance on downstream tasks. We introduce BTB3D (Better Tokens for Better 3D), a causal convolutional encoder-decoder that unifies 2D and 3D training and inference while producing compact, frequency-aware volumetric tokens. A three-stage training curriculum enables (i) local reconstruction, (ii) overlapping-window tiling, and (iii) long-context decoder refinement, during which the model learns from short slice excerpts yet generalizes to scans exceeding 300 slices without additional memory overhead. BTB3D sets a new state-of-the-art on two key tasks: it improves BLEU scores and increases clinical F1 by 40% over CT2Rep, CT-CHAT, and Merlin for report generation; and it reduces FID by 75% and halves FVD compared to GenerateCT and MedSyn for text-to-CT synthesis, producing anatomically consistent 512*512*241 volumes. These results confirm that precise three-dimensional tokenization, rather than larger language backbones alone, is essential for scalable vision-language modeling in 3D medical imaging. The codebase is available at: https://github.com/ibrahimethemhamamci/BTB3D
MCM: Mamba-based Cardiac Motion Tracking using Sequential Images in MRI
Jul 23, 2025Abstract:Myocardial motion tracking is important for assessing cardiac function and diagnosing cardiovascular diseases, for which cine cardiac magnetic resonance (CMR) has been established as the gold standard imaging modality. Many existing methods learn motion from single image pairs consisting of a reference frame and a randomly selected target frame from the cardiac cycle. However, these methods overlook the continuous nature of cardiac motion and often yield inconsistent and non-smooth motion estimations. In this work, we propose a novel Mamba-based cardiac motion tracking network (MCM) that explicitly incorporates target image sequence from the cardiac cycle to achieve smooth and temporally consistent motion tracking. By developing a bi-directional Mamba block equipped with a bi-directional scanning mechanism, our method facilitates the estimation of plausible deformation fields. With our proposed motion decoder that integrates motion information from frames adjacent to the target frame, our method further enhances temporal coherence. Moreover, by taking advantage of Mamba's structured state-space formulation, the proposed method learns the continuous dynamics of the myocardium from sequential images without increasing computational complexity. We evaluate the proposed method on two public datasets. The experimental results demonstrate that the proposed method quantitatively and qualitatively outperforms both conventional and state-of-the-art learning-based cardiac motion tracking methods. The code is available at https://github.com/yjh-0104/MCM.
CRG Score: A Distribution-Aware Clinical Metric for Radiology Report Generation
May 22, 2025Abstract:Evaluating long-context radiology report generation is challenging. NLG metrics fail to capture clinical correctness, while LLM-based metrics often lack generalizability. Clinical accuracy metrics are more relevant but are sensitive to class imbalance, frequently favoring trivial predictions. We propose the CRG Score, a distribution-aware and adaptable metric that evaluates only clinically relevant abnormalities explicitly described in reference reports. CRG supports both binary and structured labels (e.g., type, location) and can be paired with any LLM for feature extraction. By balancing penalties based on label distribution, it enables fairer, more robust evaluation and serves as a clinically aligned reward function.
Leveraging Multi-Modal Information to Enhance Dataset Distillation
May 15, 2025Abstract:Dataset distillation aims to create a compact and highly representative synthetic dataset that preserves the knowledge of a larger real dataset. While existing methods primarily focus on optimizing visual representations, incorporating additional modalities and refining object-level information can significantly improve the quality of distilled datasets. In this work, we introduce two key enhancements to dataset distillation: caption-guided supervision and object-centric masking. To integrate textual information, we propose two strategies for leveraging caption features: the feature concatenation, where caption embeddings are fused with visual features at the classification stage, and caption matching, which introduces a caption-based alignment loss during training to ensure semantic coherence between real and synthetic data. Additionally, we apply segmentation masks to isolate target objects and remove background distractions, introducing two loss functions designed for object-centric learning: masked feature alignment loss and masked gradient matching loss. Comprehensive evaluations demonstrate that integrating caption-based guidance and object-centric masking enhances dataset distillation, leading to synthetic datasets that achieve superior performance on downstream tasks.
Video Dataset Condensation with Diffusion Models
May 10, 2025Abstract:In recent years, the rapid expansion of dataset sizes and the increasing complexity of deep learning models have significantly escalated the demand for computational resources, both for data storage and model training. Dataset distillation has emerged as a promising solution to address this challenge by generating a compact synthetic dataset that retains the essential information from a large real dataset. However, existing methods often suffer from limited performance and poor data quality, particularly in the video domain. In this paper, we focus on video dataset distillation by employing a video diffusion model to generate high-quality synthetic videos. To enhance representativeness, we introduce Video Spatio-Temporal U-Net (VST-UNet), a model designed to select a diverse and informative subset of videos that effectively captures the characteristics of the original dataset. To further optimize computational efficiency, we explore a training-free clustering algorithm, Temporal-Aware Cluster-based Distillation (TAC-DT), to select representative videos without requiring additional training overhead. We validate the effectiveness of our approach through extensive experiments on four benchmark datasets, demonstrating performance improvements of up to \(10.61\%\) over the state-of-the-art. Our method consistently outperforms existing approaches across all datasets, establishing a new benchmark for video dataset distillation.
EchoFlow: A Foundation Model for Cardiac Ultrasound Image and Video Generation
Mar 28, 2025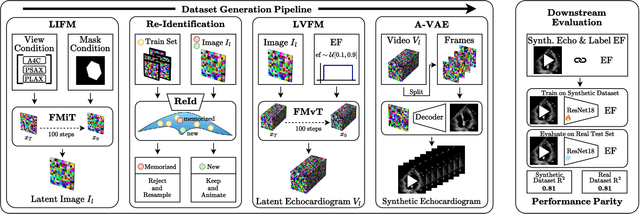
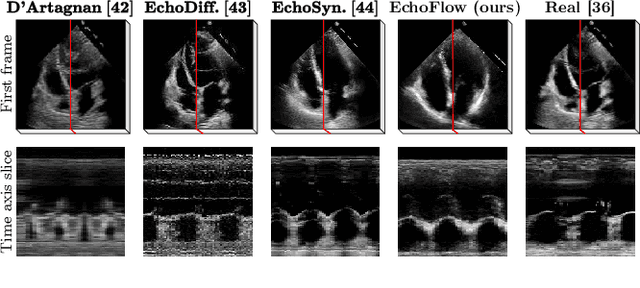
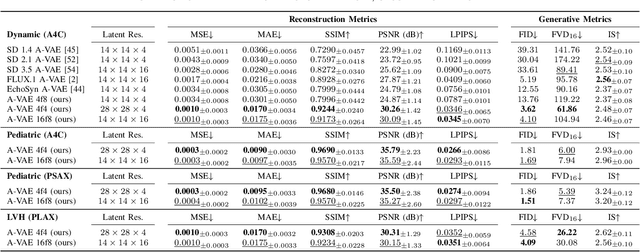
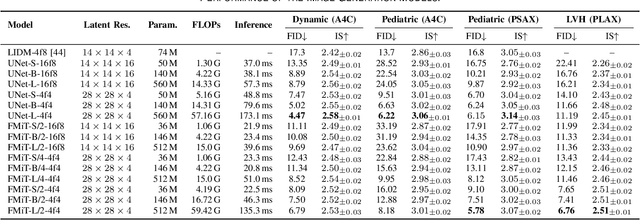
Abstract:Advances in deep learning have significantly enhanced medical image analysis, yet the availability of large-scale medical datasets remains constrained by patient privacy concerns. We present EchoFlow, a novel framework designed to generate high-quality, privacy-preserving synthetic echocardiogram images and videos. EchoFlow comprises four key components: an adversarial variational autoencoder for defining an efficient latent representation of cardiac ultrasound images, a latent image flow matching model for generating accurate latent echocardiogram images, a latent re-identification model to ensure privacy by filtering images anatomically, and a latent video flow matching model for animating latent images into realistic echocardiogram videos conditioned on ejection fraction. We rigorously evaluate our synthetic datasets on the clinically relevant task of ejection fraction regression and demonstrate, for the first time, that downstream models trained exclusively on EchoFlow-generated synthetic datasets achieve performance parity with models trained on real datasets. We release our models and synthetic datasets, enabling broader, privacy-compliant research in medical ultrasound imaging at https://huggingface.co/spaces/HReynaud/EchoFlow.
L-FUSION: Laplacian Fetal Ultrasound Segmentation & Uncertainty Estimation
Mar 07, 2025

Abstract:Accurate analysis of prenatal ultrasound (US) is essential for early detection of developmental anomalies. However, operator dependency and technical limitations (e.g. intrinsic artefacts and effects, setting errors) can complicate image interpretation and the assessment of diagnostic uncertainty. We present L-FUSION (Laplacian Fetal US Segmentation with Integrated FoundatiON models), a framework that integrates uncertainty quantification through unsupervised, normative learning and large-scale foundation models for robust segmentation of fetal structures in normal and pathological scans. We propose to utilise the aleatoric logit distributions of Stochastic Segmentation Networks and Laplace approximations with fast Hessian estimations to estimate epistemic uncertainty only from the segmentation head. This enables us to achieve reliable abnormality quantification for instant diagnostic feedback. Combined with an integrated Dropout component, L-FUSION enables reliable differentiation of lesions from normal fetal anatomy with enhanced uncertainty maps and segmentation counterfactuals in US imaging. It improves epistemic and aleatoric uncertainty interpretation and removes the need for manual disease-labelling. Evaluations across multiple datasets show that L-FUSION achieves superior segmentation accuracy and consistent uncertainty quantification, supporting on-site decision-making and offering a scalable solution for advancing fetal ultrasound analysis in clinical settings.
Image Generation Diversity Issues and How to Tame Them
Nov 25, 2024



Abstract:Generative methods now produce outputs nearly indistinguishable from real data but often fail to fully capture the data distribution. Unlike quality issues, diversity limitations in generative models are hard to detect visually, requiring specific metrics for assessment. In this paper, we draw attention to the current lack of diversity in generative models and the inability of common metrics to measure this. We achieve this by framing diversity as an image retrieval problem, where we measure how many real images can be retrieved using synthetic data as queries. This yields the Image Retrieval Score (IRS), an interpretable, hyperparameter-free metric that quantifies the diversity of a generative model's output. IRS requires only a subset of synthetic samples and provides a statistical measure of confidence. Our experiments indicate that current feature extractors commonly used in generative model assessment are inadequate for evaluating diversity effectively. Consequently, we perform an extensive search for the best feature extractors to assess diversity. Evaluation reveals that current diffusion models converge to limited subsets of the real distribution, with no current state-of-the-art models superpassing 77% of the diversity of the training data. To address this limitation, we introduce Diversity-Aware Diffusion Models (DiADM), a novel approach that improves diversity of unconditional diffusion models without loss of image quality. We do this by disentangling diversity from image quality by using a diversity aware module that uses pseudo-unconditional features as input. We provide a Python package offering unified feature extraction and metric computation to further facilitate the evaluation of generative models https://github.com/MischaD/beyondfid.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge