Jordina Aviles Verdera
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


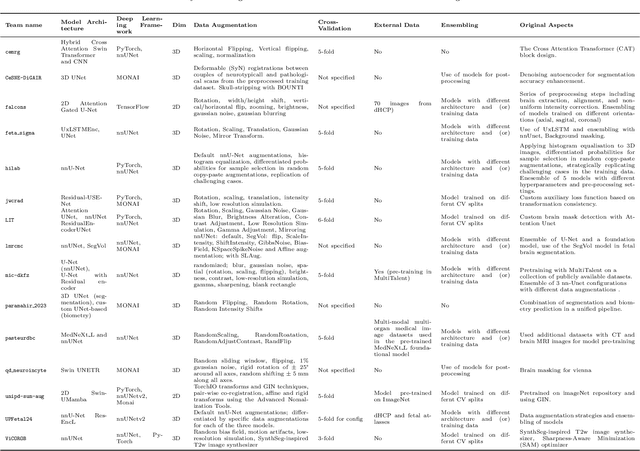
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Towards contrast- and pathology-agnostic clinical fetal brain MRI segmentation using SynthSeg
Apr 14, 2025



Abstract:Magnetic resonance imaging (MRI) has played a crucial role in fetal neurodevelopmental research. Structural annotations of MR images are an important step for quantitative analysis of the developing human brain, with Deep learning providing an automated alternative for this otherwise tedious manual process. However, segmentation performances of Convolutional Neural Networks often suffer from domain shift, where the network fails when applied to subjects that deviate from the distribution with which it is trained on. In this work, we aim to train networks capable of automatically segmenting fetal brain MRIs with a wide range of domain shifts pertaining to differences in subject physiology and acquisition environments, in particular shape-based differences commonly observed in pathological cases. We introduce a novel data-driven train-time sampling strategy that seeks to fully exploit the diversity of a given training dataset to enhance the domain generalizability of the trained networks. We adapted our sampler, together with other existing data augmentation techniques, to the SynthSeg framework, a generator that utilizes domain randomization to generate diverse training data, and ran thorough experimentations and ablation studies on a wide range of training/testing data to test the validity of the approaches. Our networks achieved notable improvements in the segmentation quality on testing subjects with intense anatomical abnormalities (p < 1e-4), though at the cost of a slighter decrease in performance in cases with fewer abnormalities. Our work also lays the foundation for future works on creating and adapting data-driven sampling strategies for other training pipelines.
Maximizing domain generalization in fetal brain tissue segmentation: the role of synthetic data generation, intensity clustering and real image fine-tuning
Nov 11, 2024



Abstract:Fetal brain tissue segmentation in magnetic resonance imaging (MRI) is a crucial tool that supports the understanding of neurodevelopment, yet it faces challenges due to the heterogeneity of data coming from different scanners and settings, and due to data scarcity. Recent approaches based on domain randomization, like SynthSeg, have shown a great potential for single source domain generalization, by simulating images with randomized contrast and image resolution from the label maps. In this work, we investigate how to maximize the out-of-domain (OOD) generalization potential of SynthSeg-based methods in fetal brain MRI. Specifically, when studying data generation, we demonstrate that the simple Gaussian mixture models used in SynthSeg enable more robust OOD generalization than physics-informed generation methods. We also investigate how intensity clustering can help create more faithful synthetic images, and observe that it is key to achieving a non-trivial OOD generalization capability when few label classes are available. Finally, by combining for the first time SynthSeg with modern fine-tuning approaches based on weight averaging, we show that fine-tuning a model pre-trained on synthetic data on a few real image-segmentation pairs in a new domain can lead to improvements in the target domain, but also in other domains. We summarize our findings as five key recommendations that we believe can guide practitioners who would like to develop SynthSeg-based approaches in other organs or modalities.
An automated pipeline for quantitative T2* fetal body MRI and segmentation at low field
Aug 09, 2023Abstract:Fetal Magnetic Resonance Imaging at low field strengths is emerging as an exciting direction in perinatal health. Clinical low field (0.55T) scanners are beneficial for fetal imaging due to their reduced susceptibility-induced artefacts, increased T2* values, and wider bore (widening access for the increasingly obese pregnant population). However, the lack of standard automated image processing tools such as segmentation and reconstruction hampers wider clinical use. In this study, we introduce a semi-automatic pipeline using quantitative MRI for the fetal body at low field strength resulting in fast and detailed quantitative T2* relaxometry analysis of all major fetal body organs. Multi-echo dynamic sequences of the fetal body were acquired and reconstructed into a single high-resolution volume using deformable slice-to-volume reconstruction, generating both structural and quantitative T2* 3D volumes. A neural network trained using a semi-supervised approach was created to automatically segment these fetal body 3D volumes into ten different organs (resulting in dice values > 0.74 for 8 out of 10 organs). The T2* values revealed a strong relationship with GA in the lungs, liver, and kidney parenchyma (R^2>0.5). This pipeline was used successfully for a wide range of GAs (17-40 weeks), and is robust to motion artefacts. Low field fetal MRI can be used to perform advanced MRI analysis, and is a viable option for clinical scanning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge