Shanshan Wang
Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, Peng Cheng Laboratory, Shenzhen, China
Semi-Supervised and Self-Supervised Collaborative Learning for Prostate 3D MR Image Segmentation
Nov 16, 2022Abstract:Volumetric magnetic resonance (MR) image segmentation plays an important role in many clinical applications. Deep learning (DL) has recently achieved state-of-the-art or even human-level performance on various image segmentation tasks. Nevertheless, manually annotating volumetric MR images for DL model training is labor-exhaustive and time-consuming. In this work, we aim to train a semi-supervised and self-supervised collaborative learning framework for prostate 3D MR image segmentation while using extremely sparse annotations, for which the ground truth annotations are provided for just the central slice of each volumetric MR image. Specifically, semi-supervised learning and self-supervised learning methods are used to generate two independent sets of pseudo labels. These pseudo labels are then fused by Boolean operation to extract a more confident pseudo label set. The images with either manual or network self-generated labels are then employed to train a segmentation model for target volume extraction. Experimental results on a publicly available prostate MR image dataset demonstrate that, while requiring significantly less annotation effort, our framework generates very encouraging segmentation results. The proposed framework is very useful in clinical applications when training data with dense annotations are difficult to obtain.
Uncertainty-Aware Multi-Parametric Magnetic Resonance Image Information Fusion for 3D Object Segmentation
Nov 16, 2022Abstract:Multi-parametric magnetic resonance (MR) imaging is an indispensable tool in the clinic. Consequently, automatic volume-of-interest segmentation based on multi-parametric MR imaging is crucial for computer-aided disease diagnosis, treatment planning, and prognosis monitoring. Despite the extensive studies conducted in deep learning-based medical image analysis, further investigations are still required to effectively exploit the information provided by different imaging parameters. How to fuse the information is a key question in this field. Here, we propose an uncertainty-aware multi-parametric MR image feature fusion method to fully exploit the information for enhanced 3D image segmentation. Uncertainties in the independent predictions of individual modalities are utilized to guide the fusion of multi-modal image features. Extensive experiments on two datasets, one for brain tissue segmentation and the other for abdominal multi-organ segmentation, have been conducted, and our proposed method achieves better segmentation performance when compared to existing models.
Adaptive PromptNet For Auxiliary Glioma Diagnosis without Contrast-Enhanced MRI
Nov 15, 2022Abstract:Multi-contrast magnetic resonance imaging (MRI)-based automatic auxiliary glioma diagnosis plays an important role in the clinic. Contrast-enhanced MRI sequences (e.g., contrast-enhanced T1-weighted imaging) were utilized in most of the existing relevant studies, in which remarkable diagnosis results have been reported. Nevertheless, acquiring contrast-enhanced MRI data is sometimes not feasible due to the patients physiological limitations. Furthermore, it is more time-consuming and costly to collect contrast-enhanced MRI data in the clinic. In this paper, we propose an adaptive PromptNet to address these issues. Specifically, a PromptNet for glioma grading utilizing only non-enhanced MRI data has been constructed. PromptNet receives constraints from features of contrast-enhanced MR data during training through a designed prompt loss. To further boost the performance, an adaptive strategy is designed to dynamically weight the prompt loss in a sample-based manner. As a result, PromptNet is capable of dealing with more difficult samples. The effectiveness of our method is evaluated on a widely-used BraTS2020 dataset, and competitive glioma grading performance on NE-MRI data is achieved.
DIGEST: Deeply supervIsed knowledGE tranSfer neTwork learning for brain tumor segmentation with incomplete multi-modal MRI scans
Nov 15, 2022Abstract:Brain tumor segmentation based on multi-modal magnetic resonance imaging (MRI) plays a pivotal role in assisting brain cancer diagnosis, treatment, and postoperative evaluations. Despite the achieved inspiring performance by existing automatic segmentation methods, multi-modal MRI data are still unavailable in real-world clinical applications due to quite a few uncontrollable factors (e.g. different imaging protocols, data corruption, and patient condition limitations), which lead to a large performance drop during practical applications. In this work, we propose a Deeply supervIsed knowledGE tranSfer neTwork (DIGEST), which achieves accurate brain tumor segmentation under different modality-missing scenarios. Specifically, a knowledge transfer learning frame is constructed, enabling a student model to learn modality-shared semantic information from a teacher model pretrained with the complete multi-modal MRI data. To simulate all the possible modality-missing conditions under the given multi-modal data, we generate incomplete multi-modal MRI samples based on Bernoulli sampling. Finally, a deeply supervised knowledge transfer loss is designed to ensure the consistency of the teacher-student structure at different decoding stages, which helps the extraction of inherent and effective modality representations. Experiments on the BraTS 2020 dataset demonstrate that our method achieves promising results for the incomplete multi-modal MR image segmentation task.
Self-supervised learning of audio representations using angular contrastive loss
Nov 10, 2022Abstract:In Self-Supervised Learning (SSL), various pretext tasks are designed for learning feature representations through contrastive loss. However, previous studies have shown that this loss is less tolerant to semantically similar samples due to the inherent defect of instance discrimination objectives, which may harm the quality of learned feature embeddings used in downstream tasks. To improve the discriminative ability of feature embeddings in SSL, we propose a new loss function called Angular Contrastive Loss (ACL), a linear combination of angular margin and contrastive loss. ACL improves contrastive learning by explicitly adding an angular margin between positive and negative augmented pairs in SSL. Experimental results show that using ACL for both supervised and unsupervised learning significantly improves performance. We validated our new loss function using the FSDnoisy18k dataset, where we achieved 73.6% and 77.1% accuracy in sound event classification using supervised and self-supervised learning, respectively.
Self-supervised Graph Learning for Long-tailed Cognitive Diagnosis
Oct 15, 2022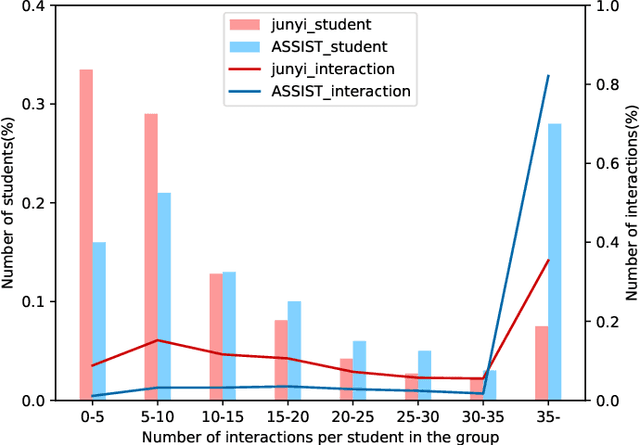
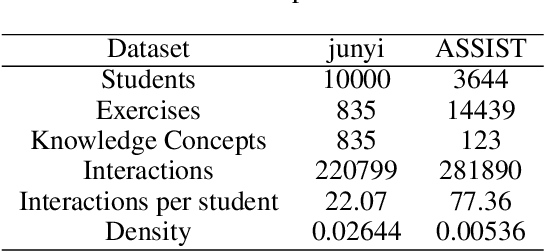
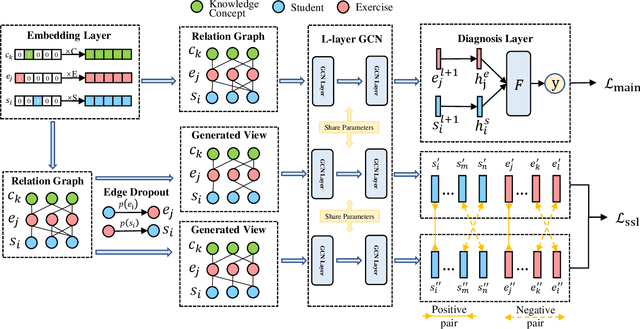
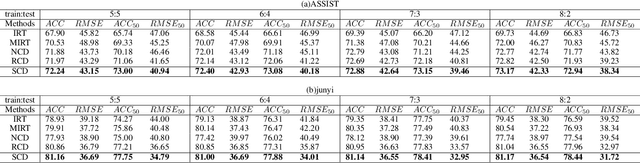
Abstract:Cognitive diagnosis is a fundamental yet critical research task in the field of intelligent education, which aims to discover the proficiency level of different students on specific knowledge concepts. Despite the effectiveness of existing efforts, previous methods always considered the mastery level on the whole students, so they still suffer from the Long Tail Effect. A large number of students who have sparse data are performed poorly in the model. To relieve the situation, we proposed a Self-supervised Cognitive Diagnosis (SCD) framework which leverages the self-supervised manner to assist the graph-based cognitive diagnosis, then the performance on those students with sparse data can be improved. Specifically, we came up with a graph confusion method that drops edges under some special rules to generate different sparse views of the graph. By maximizing the consistency of the representation on the same node under different views, the model could be more focused on long-tailed students. Additionally, we proposed an importance-based view generation rule to improve the influence of long-tailed students. Extensive experiments on real-world datasets show the effectiveness of our approach, especially on the students with sparse data.
One-shot Generative Prior Learned from Hankel-k-space for Parallel Imaging Reconstruction
Aug 28, 2022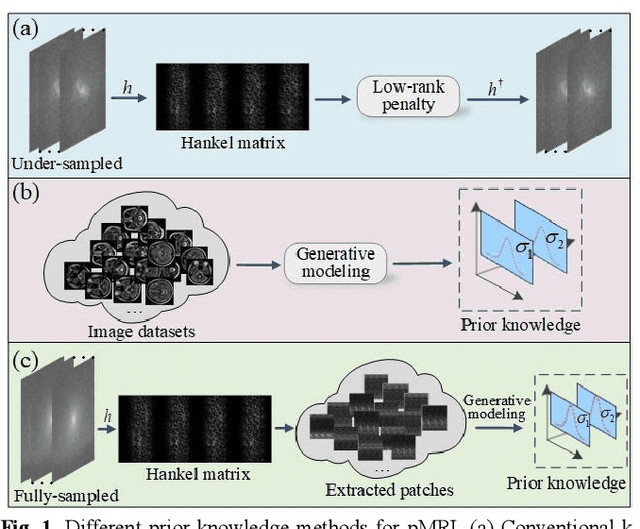
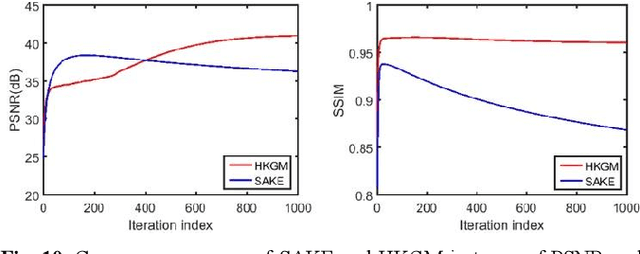
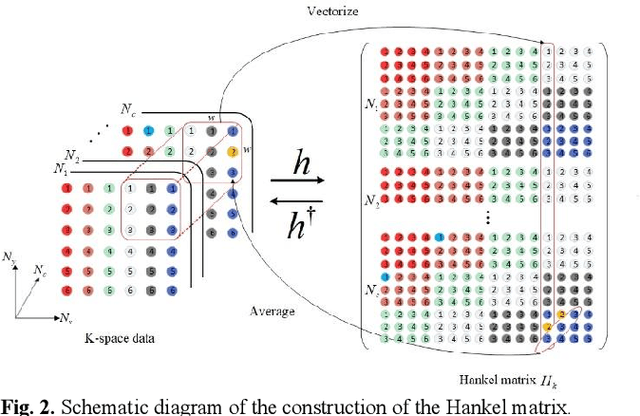
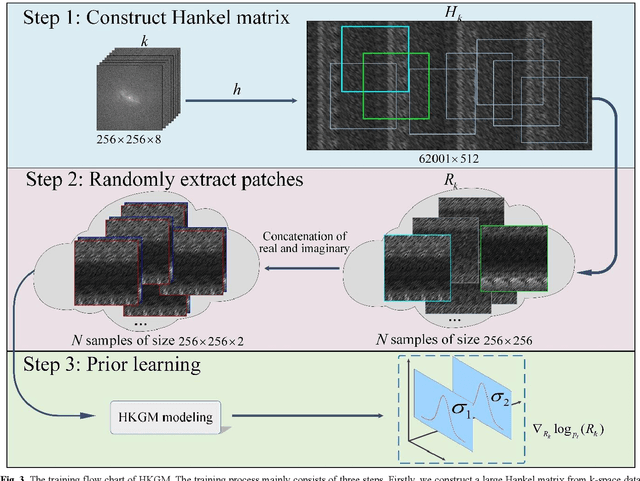
Abstract:Magnetic resonance imaging serves as an essential tool for clinical diagnosis. However, it suffers from a long acquisition time. The utilization of deep learning, especially the deep generative models, offers aggressive acceleration and better reconstruction in magnetic resonance imaging. Nevertheless, learning the data distribution as prior knowledge and reconstructing the image from limited data remains challenging. In this work, we propose a novel Hankel-k-space generative model (HKGM), which can generate samples from a training set of as little as one k-space data. At the prior learning stage, we first construct a large Hankel matrix from k-space data, then extract multiple structured k-space patches from the large Hankel matrix to capture the internal distribution among different patches. Extracting patches from a Hankel matrix enables the generative model to be learned from redundant and low-rank data space. At the iterative reconstruction stage, it is observed that the desired solution obeys the learned prior knowledge. The intermediate reconstruction solution is updated by taking it as the input of the generative model. The updated result is then alternatively operated by imposing low-rank penalty on its Hankel matrix and data consistency con-strain on the measurement data. Experimental results confirmed that the internal statistics of patches within a single k-space data carry enough information for learning a powerful generative model and provide state-of-the-art reconstruction.
SelfCoLearn: Self-supervised collaborative learning for accelerating dynamic MR imaging
Aug 08, 2022Abstract:Lately, deep learning has been extensively investigated for accelerating dynamic magnetic resonance (MR) imaging, with encouraging progresses achieved. However, without fully sampled reference data for training, current approaches may have limited abilities in recovering fine details or structures. To address this challenge, this paper proposes a self-supervised collaborative learning framework (SelfCoLearn) for accurate dynamic MR image reconstruction from undersampled k-space data. The proposed framework is equipped with three important components, namely, dual-network collaborative learning, reunderampling data augmentation and a specially designed co-training loss. The framework is flexible to be integrated with both data-driven networks and model-based iterative un-rolled networks. Our method has been evaluated on in-vivo dataset and compared it to four state-of-the-art methods. Results show that our method possesses strong capabilities in capturing essential and inherent representations for direct reconstructions from the undersampled k-space data and thus enables high-quality and fast dynamic MR imaging.
Self-supervised Learning of Audio Representations from Audio-Visual Data using Spatial Alignment
Jun 02, 2022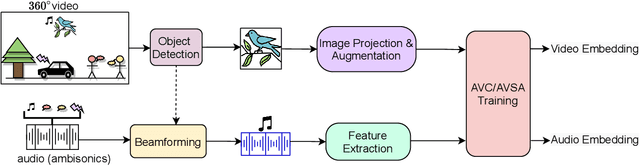
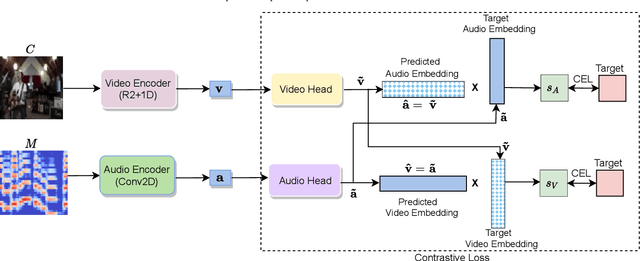
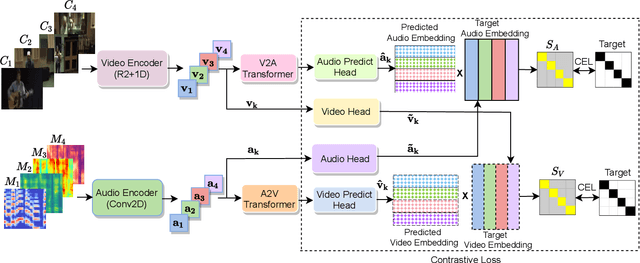
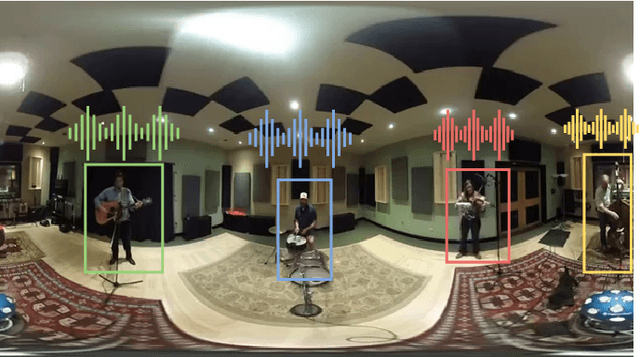
Abstract:Learning from audio-visual data offers many possibilities to express correspondence between the audio and visual content, similar to the human perception that relates aural and visual information. In this work, we present a method for self-supervised representation learning based on audio-visual spatial alignment (AVSA), a more sophisticated alignment task than the audio-visual correspondence (AVC). In addition to the correspondence, AVSA also learns from the spatial location of acoustic and visual content. Based on 360$^\text{o}$ video and Ambisonics audio, we propose selection of visual objects using object detection, and beamforming of the audio signal towards the detected objects, attempting to learn the spatial alignment between objects and the sound they produce. We investigate the use of spatial audio features to represent the audio input, and different audio formats: Ambisonics, mono, and stereo. Experimental results show a 10 $\%$ improvement on AVSA for the first order ambisonics intensity vector (FOA-IV) in comparison with log-mel spectrogram features; the addition of object-oriented crops also brings significant performance increases for the human action recognition downstream task. A number of audio-only downstream tasks are devised for testing the effectiveness of the learnt audio feature representation, obtaining performance comparable to state-of-the-art methods on acoustic scene classification from ambisonic and binaural audio.
Paying More Attention to Self-attention: Improving Pre-trained Language Models via Attention Guiding
Apr 06, 2022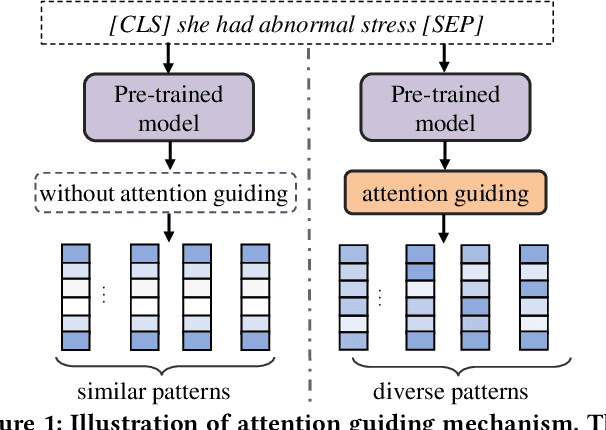
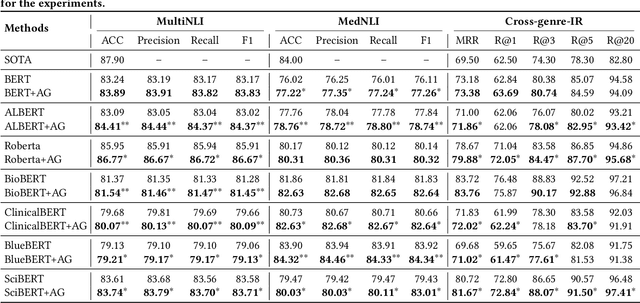
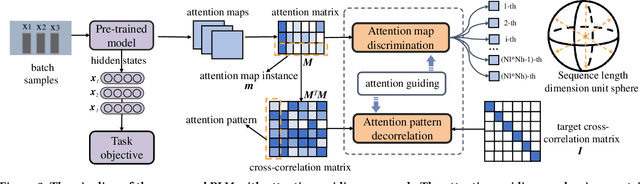
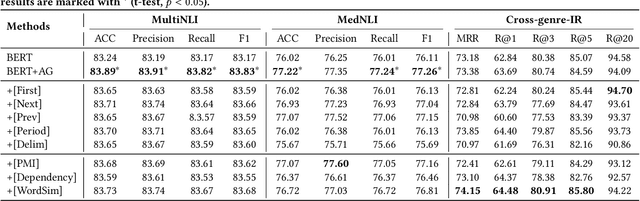
Abstract:Pre-trained language models (PLM) have demonstrated their effectiveness for a broad range of information retrieval and natural language processing tasks. As the core part of PLM, multi-head self-attention is appealing for its ability to jointly attend to information from different positions. However, researchers have found that PLM always exhibits fixed attention patterns regardless of the input (e.g., excessively paying attention to [CLS] or [SEP]), which we argue might neglect important information in the other positions. In this work, we propose a simple yet effective attention guiding mechanism to improve the performance of PLM by encouraging attention towards the established goals. Specifically, we propose two kinds of attention guiding methods, i.e., map discrimination guiding (MDG) and attention pattern decorrelation guiding (PDG). The former definitely encourages the diversity among multiple self-attention heads to jointly attend to information from different representation subspaces, while the latter encourages self-attention to attend to as many different positions of the input as possible. We conduct experiments with multiple general pre-trained models (i.e., BERT, ALBERT, and Roberta) and domain-specific pre-trained models (i.e., BioBERT, ClinicalBERT, BlueBert, and SciBERT) on three benchmark datasets (i.e., MultiNLI, MedNLI, and Cross-genre-IR). Extensive experimental results demonstrate that our proposed MDG and PDG bring stable performance improvements on all datasets with high efficiency and low cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge