Hua Han
Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, University of Chinese Academy of Sciences, Beijing, China
NeuroMamba: Multi-Perspective Feature Interaction with Visual Mamba for Neuron Segmentation
Jan 22, 2026Abstract:Neuron segmentation is the cornerstone of reconstructing comprehensive neuronal connectomes, which is essential for deciphering the functional organization of the brain. The irregular morphology and densely intertwined structures of neurons make this task particularly challenging. Prevailing CNN-based methods often fail to resolve ambiguous boundaries due to the lack of long-range context, whereas Transformer-based methods suffer from boundary imprecision caused by the loss of voxel-level details during patch partitioning. To address these limitations, we propose NeuroMamba, a multi-perspective framework that exploits the linear complexity of Mamba to enable patch-free global modeling and synergizes this with complementary local feature modeling, thereby efficiently capturing long-range dependencies while meticulously preserving fine-grained voxel details. Specifically, we design a channel-gated Boundary Discriminative Feature Extractor (BDFE) to enhance local morphological cues. Complementing this, we introduce the Spatial Continuous Feature Extractor (SCFE), which integrates a resolution-aware scanning mechanism into the Visual Mamba architecture to adaptively model global dependencies across varying data resolutions. Finally, a cross-modulation mechanism synergistically fuses these multi-perspective features. Our method demonstrates state-of-the-art performance across four public EM datasets, validating its exceptional adaptability to both anisotropic and isotropic resolutions. The source code will be made publicly available.
Diffusion Model-Based Data Augmentation for Enhanced Neuron Segmentation
Jan 22, 2026Abstract:Neuron segmentation in electron microscopy (EM) aims to reconstruct the complete neuronal connectome; however, current deep learning-based methods are limited by their reliance on large-scale training data and extensive, time-consuming manual annotations. Traditional methods augment the training set through geometric and photometric transformations; however, the generated samples remain highly correlated with the original images and lack structural diversity. To address this limitation, we propose a diffusion-based data augmentation framework capable of generating diverse and structurally plausible image-label pairs for neuron segmentation. Specifically, the framework employs a resolution-aware conditional diffusion model with multi-scale conditioning and EM resolution priors to enable voxel-level image synthesis from 3D masks. It further incorporates a biology-guided mask remodeling module that produces augmented masks with enhanced structural realism. Together, these components effectively enrich the training set and improve segmentation performance. On the AC3 and AC4 datasets under low-annotation regimes, our method improves the ARAND metric by 32.1% and 30.7%, respectively, when combined with two different post-processing methods. Our code is available at https://github.com/HeadLiuYun/NeuroDiff.
From Diffusion to Resolution: Leveraging 2D Diffusion Models for 3D Super-Resolution Task
Nov 25, 2024



Abstract:Diffusion models have recently emerged as a powerful technique in image generation, especially for image super-resolution tasks. While 2D diffusion models significantly enhance the resolution of individual images, existing diffusion-based methods for 3D volume super-resolution often struggle with structure discontinuities in axial direction and high sampling costs. In this work, we present a novel approach that leverages the 2D diffusion model and lateral continuity within the volume to enhance 3D volume electron microscopy (vEM) super-resolution. We first simulate lateral degradation with slices in the XY plane and train a 2D diffusion model to learn how to restore the degraded slices. The model is then applied slice-by-slice in the lateral direction of low-resolution volume, recovering slices while preserving inherent lateral continuity. Following this, a high-frequency-aware 3D super-resolution network is trained on the recovery lateral slice sequences to learn spatial feature transformation across slices. Finally, the network is applied to infer high-resolution volumes in the axial direction, enabling 3D super-resolution. We validate our approach through comprehensive evaluations, including image similarity assessments, resolution analysis, and performance on downstream tasks. Our results on two publicly available focused ion beam scanning electron microscopy (FIB-SEM) datasets demonstrate the robustness and practical applicability of our framework for 3D volume super-resolution.
LESEN: Label-Efficient deep learning for Multi-parametric MRI-based Visual Pathway Segmentation
Jan 03, 2024Abstract:Recent research has shown the potential of deep learning in multi-parametric MRI-based visual pathway (VP) segmentation. However, obtaining labeled data for training is laborious and time-consuming. Therefore, it is crucial to develop effective algorithms in situations with limited labeled samples. In this work, we propose a label-efficient deep learning method with self-ensembling (LESEN). LESEN incorporates supervised and unsupervised losses, enabling the student and teacher models to mutually learn from each other, forming a self-ensembling mean teacher framework. Additionally, we introduce a reliable unlabeled sample selection (RUSS) mechanism to further enhance LESEN's effectiveness. Our experiments on the human connectome project (HCP) dataset demonstrate the superior performance of our method when compared to state-of-the-art techniques, advancing multimodal VP segmentation for comprehensive analysis in clinical and research settings. The implementation code will be available at: https://github.com/aldiak/Semi-Supervised-Multimodal-Visual-Pathway- Delineation.
Modality Exchange Network for Retinogeniculate Visual Pathway Segmentation
Jan 03, 2024Abstract:Accurate segmentation of the retinogeniculate visual pathway (RGVP) aids in the diagnosis and treatment of visual disorders by identifying disruptions or abnormalities within the pathway. However, the complex anatomical structure and connectivity of RGVP make it challenging to achieve accurate segmentation. In this study, we propose a novel Modality Exchange Network (ME-Net) that effectively utilizes multi-modal magnetic resonance (MR) imaging information to enhance RGVP segmentation. Our ME-Net has two main contributions. Firstly, we introduce an effective multi-modal soft-exchange technique. Specifically, we design a channel and spatially mixed attention module to exchange modality information between T1-weighted and fractional anisotropy MR images. Secondly, we propose a cross-fusion module that further enhances the fusion of information between the two modalities. Experimental results demonstrate that our method outperforms existing state-of-the-art approaches in terms of RGVP segmentation performance.
Blind2Sound: Self-Supervised Image Denoising without Residual Noise
Mar 14, 2023



Abstract:Self-supervised blind denoising for Poisson-Gaussian noise remains a challenging task. Pseudo-supervised pairs constructed from single noisy images re-corrupt the signal and degrade the performance. The visible blindspots solve the information loss in masked inputs. However, without explicitly noise sensing, mean square error as an objective function cannot adjust denoising intensities for dynamic noise levels, leading to noticeable residual noise. In this paper, we propose Blind2Sound, a simple yet effective approach to overcome residual noise in denoised images. The proposed adaptive re-visible loss senses noise levels and performs personalized denoising without noise residues while retaining the signal lossless. The theoretical analysis of intermediate medium gradients guarantees stable training, while the Cramer Gaussian loss acts as a regularization to facilitate the accurate perception of noise levels and improve the performance of the denoiser. Experiments on synthetic and real-world datasets show the superior performance of our method, especially for single-channel images.
Uncertainty-Aware Multi-Parametric Magnetic Resonance Image Information Fusion for 3D Object Segmentation
Nov 16, 2022



Abstract:Multi-parametric magnetic resonance (MR) imaging is an indispensable tool in the clinic. Consequently, automatic volume-of-interest segmentation based on multi-parametric MR imaging is crucial for computer-aided disease diagnosis, treatment planning, and prognosis monitoring. Despite the extensive studies conducted in deep learning-based medical image analysis, further investigations are still required to effectively exploit the information provided by different imaging parameters. How to fuse the information is a key question in this field. Here, we propose an uncertainty-aware multi-parametric MR image feature fusion method to fully exploit the information for enhanced 3D image segmentation. Uncertainties in the independent predictions of individual modalities are utilized to guide the fusion of multi-modal image features. Extensive experiments on two datasets, one for brain tissue segmentation and the other for abdominal multi-organ segmentation, have been conducted, and our proposed method achieves better segmentation performance when compared to existing models.
Blind2Unblind: Self-Supervised Image Denoising with Visible Blind Spots
Mar 15, 2022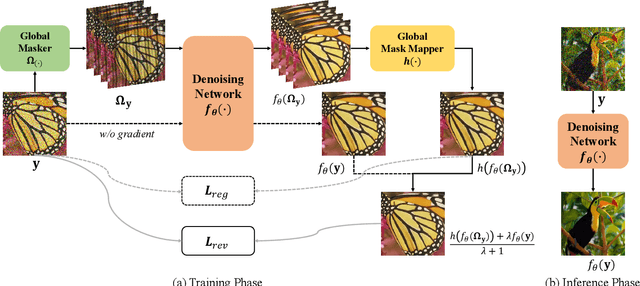
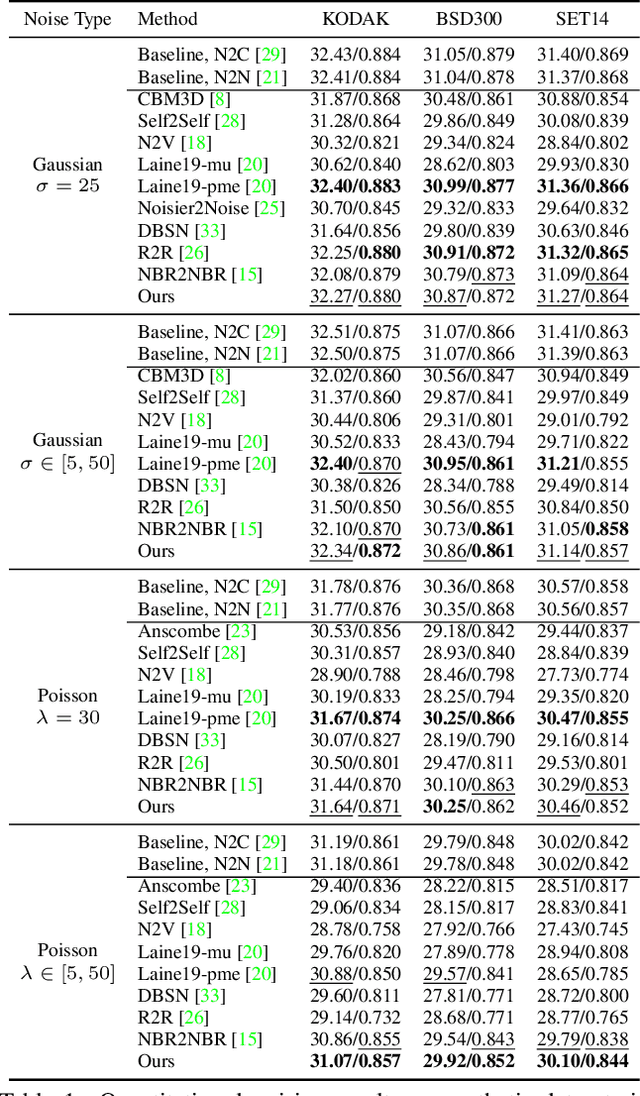
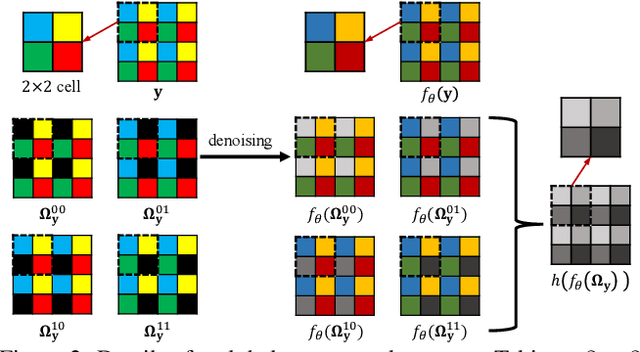
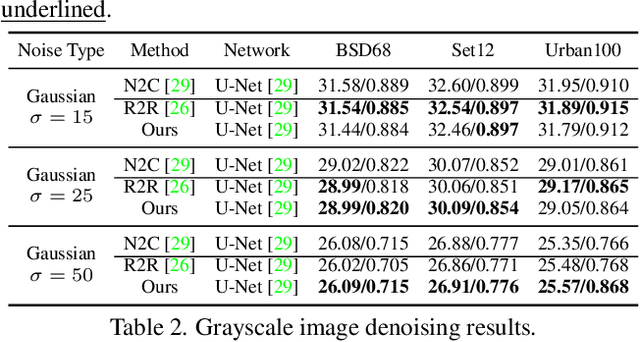
Abstract:Real noisy-clean pairs on a large scale are costly and difficult to obtain. Meanwhile, supervised denoisers trained on synthetic data perform poorly in practice. Self-supervised denoisers, which learn only from single noisy images, solve the data collection problem. However, self-supervised denoising methods, especially blindspot-driven ones, suffer sizable information loss during input or network design. The absence of valuable information dramatically reduces the upper bound of denoising performance. In this paper, we propose a simple yet efficient approach called Blind2Unblind to overcome the information loss in blindspot-driven denoising methods. First, we introduce a global-aware mask mapper that enables global perception and accelerates training. The mask mapper samples all pixels at blind spots on denoised volumes and maps them to the same channel, allowing the loss function to optimize all blind spots at once. Second, we propose a re-visible loss to train the denoising network and make blind spots visible. The denoiser can learn directly from raw noise images without losing information or being trapped in identity mapping. We also theoretically analyze the convergence of the re-visible loss. Extensive experiments on synthetic and real-world datasets demonstrate the superior performance of our approach compared to previous work. Code is available at https://github.com/demonsjin/Blind2Unblind.
Temporal Spatial-Adaptive Interpolation with Deformable Refinement for Electron Microscopic Images
Jan 17, 2021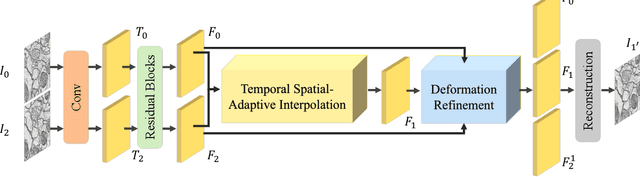
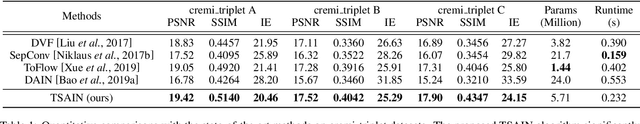
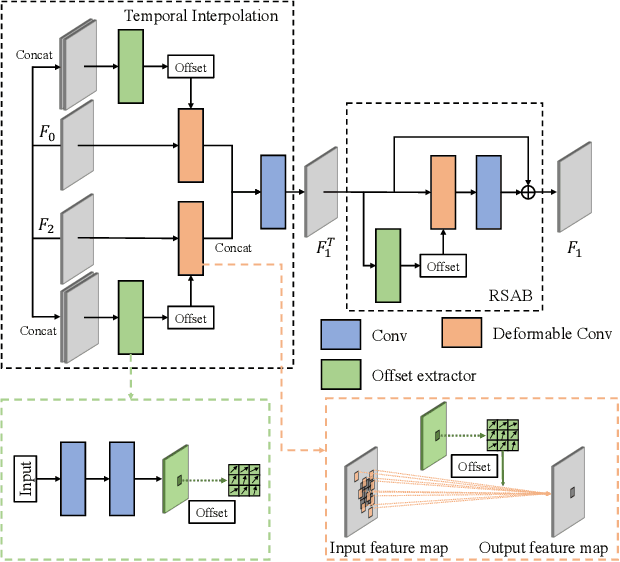
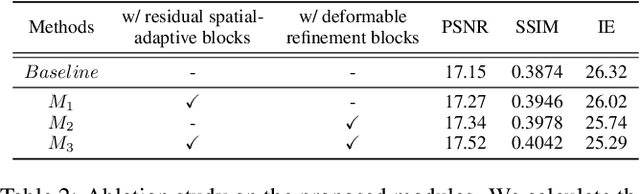
Abstract:Recently, flow-based methods have achieved promising success in video frame interpolation. However, electron microscopic (EM) images suffer from unstable image quality, low PSNR, and disorderly deformation. Existing flow-based interpolation methods cannot precisely compute optical flow for EM images since only predicting each position's unique offset. To overcome these problems, we propose a novel interpolation framework for EM images that progressively synthesizes interpolated features in a coarse-to-fine manner. First, we extract missing intermediate features by the proposed temporal spatial-adaptive (TSA) interpolation module. The TSA interpolation module aggregates temporal contexts and then adaptively samples the spatial-related features with the proposed residual spatial adaptive block. Second, we introduce a stacked deformable refinement block (SDRB) further enhance the reconstruction quality, which is aware of the matching positions and relevant features from input frames with the feedback mechanism. Experimental results demonstrate the superior performance of our approach compared to previous works, both quantitatively and qualitatively.
Non-iterative Simultaneous Rigid Registration Method for Serial Sections of Biological Tissue
May 11, 2020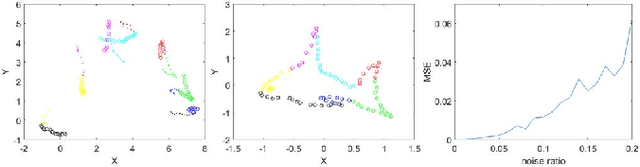
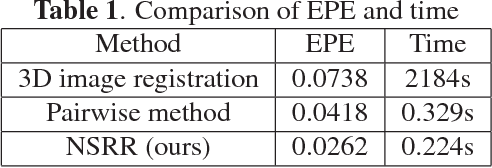
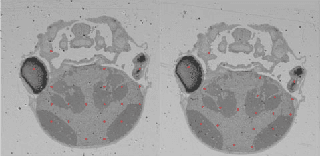
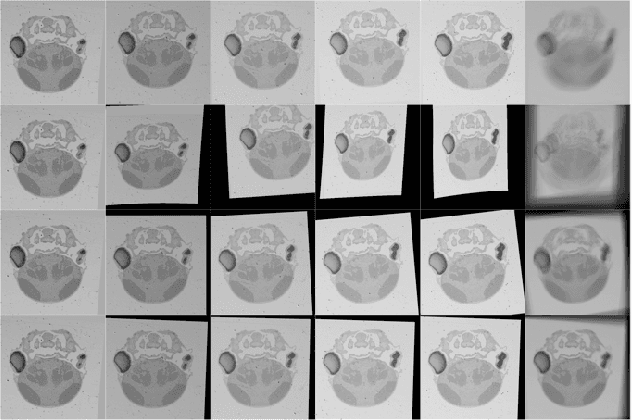
Abstract:In this paper, we propose a novel non-iterative algorithm to simultaneously estimate optimal rigid transformation for serial section images, which is a key component in volume reconstruction of serial sections of biological tissue. In order to avoid error accumulation and propagation caused by current algorithms, we add extra condition that the position of the first and the last section images should remain unchanged. This constrained simultaneous registration problem has not been solved before. Our algorithm method is non-iterative, it can simultaneously compute rigid transformation for a large number of serial section images in a short time. We prove that our algorithm gets optimal solution under ideal condition. And we test our algorithm with synthetic data and real data to verify our algorithm's effectiveness.
* appears in IEEE International Symposium on Biomedical Imaging 2018 (ISBI 2018)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge