Saumya Gupta
DeepWeightFlow: Re-Basined Flow Matching for Generating Neural Network Weights
Jan 08, 2026Abstract:Building efficient and effective generative models for neural network weights has been a research focus of significant interest that faces challenges posed by the high-dimensional weight spaces of modern neural networks and their symmetries. Several prior generative models are limited to generating partial neural network weights, particularly for larger models, such as ResNet and ViT. Those that do generate complete weights struggle with generation speed or require finetuning of the generated models. In this work, we present DeepWeightFlow, a Flow Matching model that operates directly in weight space to generate diverse and high-accuracy neural network weights for a variety of architectures, neural network sizes, and data modalities. The neural networks generated by DeepWeightFlow do not require fine-tuning to perform well and can scale to large networks. We apply Git Re-Basin and TransFusion for neural network canonicalization in the context of generative weight models to account for the impact of neural network permutation symmetries and to improve generation efficiency for larger model sizes. The generated networks excel at transfer learning, and ensembles of hundreds of neural networks can be generated in minutes, far exceeding the efficiency of diffusion-based methods. DeepWeightFlow models pave the way for more efficient and scalable generation of diverse sets of neural networks.
Image Synthesis Using Spintronic Deep Convolutional Generative Adversarial Network
Jan 04, 2026Abstract:The computational requirements of generative adversarial networks (GANs) exceed the limit of conventional Von Neumann architectures, necessitating energy efficient alternatives such as neuromorphic spintronics. This work presents a hybrid CMOS-spintronic deep convolutional generative adversarial network (DCGAN) architecture for synthetic image generation. The proposed generative vision model approach follows the standard framework, leveraging generator and discriminators adversarial training with our designed spintronics hardware for deconvolution, convolution, and activation layers of the DCGAN architecture. To enable hardware aware spintronic implementation, the generator's deconvolution layers are restructured as zero padded convolution, allowing seamless integration with a 6-bit skyrmion based synapse in a crossbar, without compromising training performance. Nonlinear activation functions are implemented using a hybrid CMOS domain wall based Rectified linear unit (ReLU) and Leaky ReLU units. Our proposed tunable Leaky ReLU employs domain wall position coded, continuous resistance states and a piecewise uniaxial parabolic anisotropy profile with a parallel MTJ readout, exhibiting energy consumption of 0.192 pJ. Our spintronic DCGAN model demonstrates adaptability across both grayscale and colored datasets, achieving Fr'echet Inception Distances (FID) of 27.5 for the Fashion MNIST and 45.4 for Anime Face datasets, with testing energy (training energy) of 4.9 nJ (14.97~nJ/image) and 24.72 nJ (74.7 nJ/image).
BrainMRDiff: A Diffusion Model for Anatomically Consistent Brain MRI Synthesis
Apr 06, 2025



Abstract:Accurate brain tumor diagnosis relies on the assessment of multiple Magnetic Resonance Imaging (MRI) sequences. However, in clinical practice, the acquisition of certain sequences may be affected by factors like motion artifacts or contrast agent contraindications, leading to suboptimal outcome, such as poor image quality. This can then affect image interpretation by radiologists. Synthesizing high quality MRI sequences has thus become a critical research focus. Though recent advancements in controllable generative AI have facilitated the synthesis of diagnostic quality MRI, ensuring anatomical accuracy remains a significant challenge. Preserving critical structural relationships between different anatomical regions is essential, as even minor structural or topological inconsistencies can compromise diagnostic validity. In this work, we propose BrainMRDiff, a novel topology-preserving, anatomy-guided diffusion model for synthesizing brain MRI, leveraging brain and tumor anatomies as conditioning inputs. To achieve this, we introduce two key modules: Tumor+Structure Aggregation (TSA) and Topology-Guided Anatomy Preservation (TGAP). TSA integrates diverse anatomical structures with tumor information, forming a comprehensive conditioning mechanism for the diffusion process. TGAP enforces topological consistency during reverse denoising diffusion process; both these modules ensure that the generated image respects anatomical integrity. Experimental results demonstrate that BrainMRDiff surpasses existing baselines, achieving performance improvements of 23.33% on the BraTS-AG dataset and 33.33% on the BraTS-Met dataset. Code will be made publicly available soon.
TopoCellGen: Generating Histopathology Cell Topology with a Diffusion Model
Dec 08, 2024Abstract:Accurately modeling multi-class cell topology is crucial in digital pathology, as it provides critical insights into tissue structure and pathology. The synthetic generation of cell topology enables realistic simulations of complex tissue environments, enhances downstream tasks by augmenting training data, aligns more closely with pathologists' domain knowledge, and offers new opportunities for controlling and generalizing the tumor microenvironment. In this paper, we propose a novel approach that integrates topological constraints into a diffusion model to improve the generation of realistic, contextually accurate cell topologies. Our method refines the simulation of cell distributions and interactions, increasing the precision and interpretability of results in downstream tasks such as cell detection and classification. To assess the topological fidelity of generated layouts, we introduce a new metric, Topological Frechet Distance (TopoFD), which overcomes the limitations of traditional metrics like FID in evaluating topological structure. Experimental results demonstrate the effectiveness of our approach in generating multi-class cell layouts that capture intricate topological relationships.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
TopoDiffusionNet: A Topology-aware Diffusion Model
Oct 22, 2024Abstract:Diffusion models excel at creating visually impressive images but often struggle to generate images with a specified topology. The Betti number, which represents the number of structures in an image, is a fundamental measure in topology. Yet, diffusion models fail to satisfy even this basic constraint. This limitation restricts their utility in applications requiring exact control, like robotics and environmental modeling. To address this, we propose TopoDiffusionNet (TDN), a novel approach that enforces diffusion models to maintain the desired topology. We leverage tools from topological data analysis, particularly persistent homology, to extract the topological structures within an image. We then design a topology-based objective function to guide the denoising process, preserving intended structures while suppressing noisy ones. Our experiments across four datasets demonstrate significant improvements in topological accuracy. TDN is the first to integrate topology with diffusion models, opening new avenues of research in this area.
Backdooring Vision-Language Models with Out-Of-Distribution Data
Oct 02, 2024



Abstract:The emergence of Vision-Language Models (VLMs) represents a significant advancement in integrating computer vision with Large Language Models (LLMs) to generate detailed text descriptions from visual inputs. Despite their growing importance, the security of VLMs, particularly against backdoor attacks, is under explored. Moreover, prior works often assume attackers have access to the original training data, which is often unrealistic. In this paper, we address a more practical and challenging scenario where attackers must rely solely on Out-Of-Distribution (OOD) data. We introduce VLOOD (Backdooring Vision-Language Models with Out-of-Distribution Data), a novel approach with two key contributions: (1) demonstrating backdoor attacks on VLMs in complex image-to-text tasks while minimizing degradation of the original semantics under poisoned inputs, and (2) proposing innovative techniques for backdoor injection without requiring any access to the original training data. Our evaluation on image captioning and visual question answering (VQA) tasks confirms the effectiveness of VLOOD, revealing a critical security vulnerability in VLMs and laying the foundation for future research on securing multimodal models against sophisticated threats.
TopoSemiSeg: Enforcing Topological Consistency for Semi-Supervised Segmentation of Histopathology Images
Dec 06, 2023Abstract:In computational pathology, segmenting densely distributed objects like glands and nuclei is crucial for downstream analysis. To alleviate the burden of obtaining pixel-wise annotations, semi-supervised learning methods learn from large amounts of unlabeled data. Nevertheless, existing semi-supervised methods overlook the topological information hidden in the unlabeled images and are thus prone to topological errors, e.g., missing or incorrectly merged/separated glands or nuclei. To address this issue, we propose TopoSemiSeg, the first semi-supervised method that learns the topological representation from unlabeled data. In particular, we propose a topology-aware teacher-student approach in which the teacher and student networks learn shared topological representations. To achieve this, we introduce topological consistency loss, which contains signal consistency and noise removal losses to ensure the learned representation is robust and focuses on true topological signals. Extensive experiments on public pathology image datasets show the superiority of our method, especially on topology-wise evaluation metrics. Code is available at https://github.com/Melon-Xu/TopoSemiSeg.
Topology-Aware Uncertainty for Image Segmentation
Jun 09, 2023



Abstract:Segmentation of curvilinear structures such as vasculature and road networks is challenging due to relatively weak signals and complex geometry/topology. To facilitate and accelerate large scale annotation, one has to adopt semi-automatic approaches such as proofreading by experts. In this work, we focus on uncertainty estimation for such tasks, so that highly uncertain, and thus error-prone structures can be identified for human annotators to verify. Unlike most existing works, which provide pixel-wise uncertainty maps, we stipulate it is crucial to estimate uncertainty in the units of topological structures, e.g., small pieces of connections and branches. To achieve this, we leverage tools from topological data analysis, specifically discrete Morse theory (DMT), to first capture the structures, and then reason about their uncertainties. To model the uncertainty, we (1) propose a joint prediction model that estimates the uncertainty of a structure while taking the neighboring structures into consideration (inter-structural uncertainty); (2) propose a novel Probabilistic DMT to model the inherent uncertainty within each structure (intra-structural uncertainty) by sampling its representations via a perturb-and-walk scheme. On various 2D and 3D datasets, our method produces better structure-wise uncertainty maps compared to existing works.
Learning Topological Interactions for Multi-Class Medical Image Segmentation
Jul 20, 2022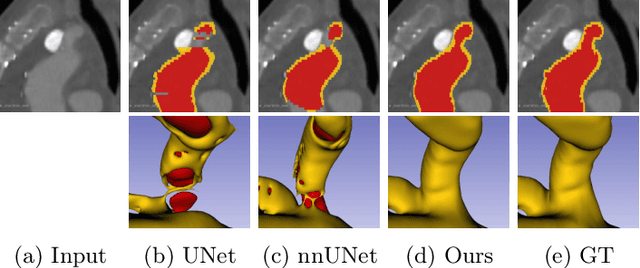
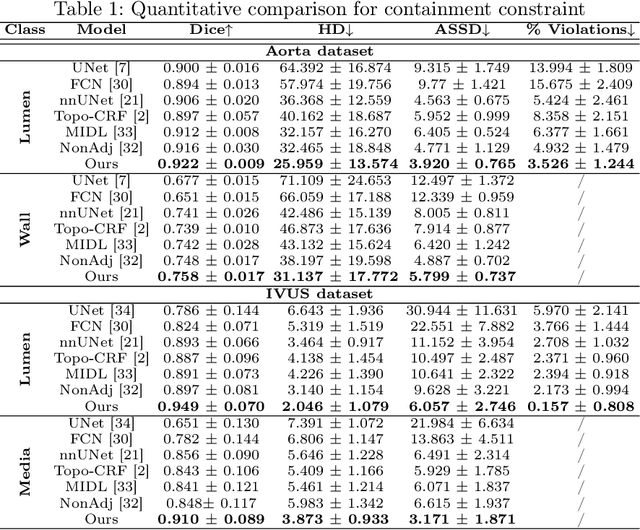
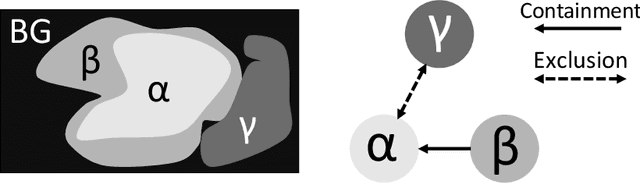
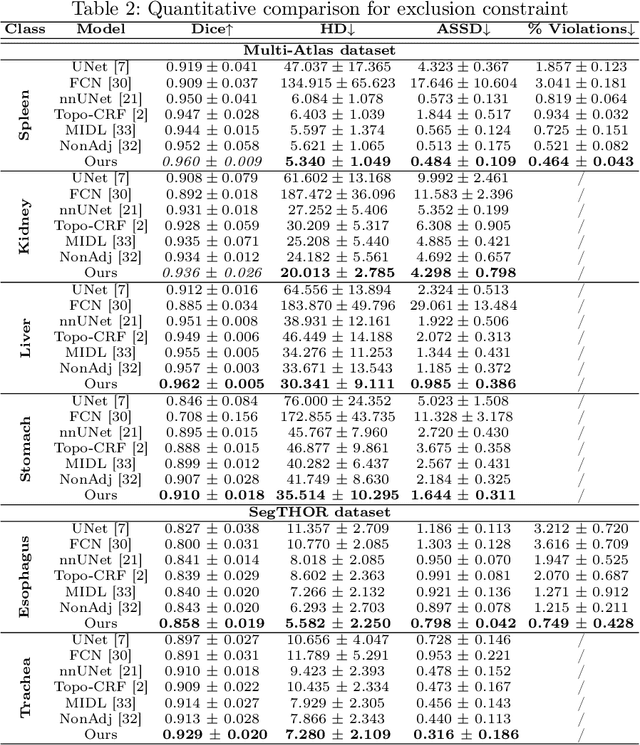
Abstract:Deep learning methods have achieved impressive performance for multi-class medical image segmentation. However, they are limited in their ability to encode topological interactions among different classes (e.g., containment and exclusion). These constraints naturally arise in biomedical images and can be crucial in improving segmentation quality. In this paper, we introduce a novel topological interaction module to encode the topological interactions into a deep neural network. The implementation is completely convolution-based and thus can be very efficient. This empowers us to incorporate the constraints into end-to-end training and enrich the feature representation of neural networks. The efficacy of the proposed method is validated on different types of interactions. We also demonstrate the generalizability of the method on both proprietary and public challenge datasets, in both 2D and 3D settings, as well as across different modalities such as CT and Ultrasound. Code is available at: https://github.com/TopoXLab/TopoInteraction
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge