Dimitris Samaras
SUNY
PISCES: Annotation-free Text-to-Video Post-Training via Optimal Transport-Aligned Rewards
Feb 02, 2026Abstract:Text-to-video (T2V) generation aims to synthesize videos with high visual quality and temporal consistency that are semantically aligned with input text. Reward-based post-training has emerged as a promising direction to improve the quality and semantic alignment of generated videos. However, recent methods either rely on large-scale human preference annotations or operate on misaligned embeddings from pre-trained vision-language models, leading to limited scalability or suboptimal supervision. We present $\texttt{PISCES}$, an annotation-free post-training algorithm that addresses these limitations via a novel Dual Optimal Transport (OT)-aligned Rewards module. To align reward signals with human judgment, $\texttt{PISCES}$ uses OT to bridge text and video embeddings at both distributional and discrete token levels, enabling reward supervision to fulfill two objectives: (i) a Distributional OT-aligned Quality Reward that captures overall visual quality and temporal coherence; and (ii) a Discrete Token-level OT-aligned Semantic Reward that enforces semantic, spatio-temporal correspondence between text and video tokens. To our knowledge, $\texttt{PISCES}$ is the first to improve annotation-free reward supervision in generative post-training through the lens of OT. Experiments on both short- and long-video generation show that $\texttt{PISCES}$ outperforms both annotation-based and annotation-free methods on VBench across Quality and Semantic scores, with human preference studies further validating its effectiveness. We show that the Dual OT-aligned Rewards module is compatible with multiple optimization paradigms, including direct backpropagation and reinforcement learning fine-tuning.
LVLMs and Humans Ground Differently in Referential Communication
Jan 28, 2026Abstract:For generative AI agents to partner effectively with human users, the ability to accurately predict human intent is critical. But this ability to collaborate remains limited by a critical deficit: an inability to model common ground. Here, we present a referential communication experiment with a factorial design involving director-matcher pairs (human-human, human-AI, AI-human, and AI-AI) that interact with multiple turns in repeated rounds to match pictures of objects not associated with any obvious lexicalized labels. We release the online pipeline for data collection, the tools and analyses for accuracy, efficiency, and lexical overlap, and a corpus of 356 dialogues (89 pairs over 4 rounds each) that unmasks LVLMs' limitations in interactively resolving referring expressions, a crucial skill that underlies human language use.
Generating metamers of human scene understanding
Jan 16, 2026Abstract:Human vision combines low-resolution "gist" information from the visual periphery with sparse but high-resolution information from fixated locations to construct a coherent understanding of a visual scene. In this paper, we introduce MetamerGen, a tool for generating scenes that are aligned with latent human scene representations. MetamerGen is a latent diffusion model that combines peripherally obtained scene gist information with information obtained from scene-viewing fixations to generate image metamers for what humans understand after viewing a scene. Generating images from both high and low resolution (i.e. "foveated") inputs constitutes a novel image-to-image synthesis problem, which we tackle by introducing a dual-stream representation of the foveated scenes consisting of DINOv2 tokens that fuse detailed features from fixated areas with peripherally degraded features capturing scene context. To evaluate the perceptual alignment of MetamerGen generated images to latent human scene representations, we conducted a same-different behavioral experiment where participants were asked for a "same" or "different" response between the generated and the original image. With that, we identify scene generations that are indeed metamers for the latent scene representations formed by the viewers. MetamerGen is a powerful tool for understanding scene understanding. Our proof-of-concept analyses uncovered specific features at multiple levels of visual processing that contributed to human judgments. While it can generate metamers even conditioned on random fixations, we find that high-level semantic alignment most strongly predicts metamerism when the generated scenes are conditioned on viewers' own fixated regions.
GriDiT: Factorized Grid-Based Diffusion for Efficient Long Image Sequence Generation
Dec 24, 2025



Abstract:Modern deep learning methods typically treat image sequences as large tensors of sequentially stacked frames. However, is this straightforward representation ideal given the current state-of-the-art (SoTA)? In this work, we address this question in the context of generative models and aim to devise a more effective way of modeling image sequence data. Observing the inefficiencies and bottlenecks of current SoTA image sequence generation methods, we showcase that rather than working with large tensors, we can improve the generation process by factorizing it into first generating the coarse sequence at low resolution and then refining the individual frames at high resolution. We train a generative model solely on grid images comprising subsampled frames. Yet, we learn to generate image sequences, using the strong self-attention mechanism of the Diffusion Transformer (DiT) to capture correlations between frames. In effect, our formulation extends a 2D image generator to operate as a low-resolution 3D image-sequence generator without introducing any architectural modifications. Subsequently, we super-resolve each frame individually to add the sequence-independent high-resolution details. This approach offers several advantages and can overcome key limitations of the SoTA in this domain. Compared to existing image sequence generation models, our method achieves superior synthesis quality and improved coherence across sequences. It also delivers high-fidelity generation of arbitrary-length sequences and increased efficiency in inference time and training data usage. Furthermore, our straightforward formulation enables our method to generalize effectively across diverse data domains, which typically require additional priors and supervision to model in a generative context. Our method consistently outperforms SoTA in quality and inference speed (at least twice-as-fast) across datasets.
TICON: A Slide-Level Tile Contextualizer for Histopathology Representation Learning
Dec 24, 2025Abstract:The interpretation of small tiles in large whole slide images (WSI) often needs a larger image context. We introduce TICON, a transformer-based tile representation contextualizer that produces rich, contextualized embeddings for ''any'' application in computational pathology. Standard tile encoder-based pipelines, which extract embeddings of tiles stripped from their context, fail to model the rich slide-level information essential for both local and global tasks. Furthermore, different tile-encoders excel at different downstream tasks. Therefore, a unified model is needed to contextualize embeddings derived from ''any'' tile-level foundation model. TICON addresses this need with a single, shared encoder, pretrained using a masked modeling objective to simultaneously unify and contextualize representations from diverse tile-level pathology foundation models. Our experiments demonstrate that TICON-contextualized embeddings significantly improve performance across many different tasks, establishing new state-of-the-art results on tile-level benchmarks (i.e., HEST-Bench, THUNDER, CATCH) and slide-level benchmarks (i.e., Patho-Bench). Finally, we pretrain an aggregator on TICON to form a slide-level foundation model, using only 11K WSIs, outperforming SoTA slide-level foundation models pretrained with up to 350K WSIs.
PixCell: A generative foundation model for digital histopathology images
Jun 05, 2025Abstract:The digitization of histology slides has revolutionized pathology, providing massive datasets for cancer diagnosis and research. Contrastive self-supervised and vision-language models have been shown to effectively mine large pathology datasets to learn discriminative representations. On the other hand, generative models, capable of synthesizing realistic and diverse images, present a compelling solution to address unique problems in pathology that involve synthesizing images; overcoming annotated data scarcity, enabling privacy-preserving data sharing, and performing inherently generative tasks, such as virtual staining. We introduce PixCell, the first diffusion-based generative foundation model for histopathology. We train PixCell on PanCan-30M, a vast, diverse dataset derived from 69,184 H\&E-stained whole slide images covering various cancer types. We employ a progressive training strategy and a self-supervision-based conditioning that allows us to scale up training without any annotated data. PixCell generates diverse and high-quality images across multiple cancer types, which we find can be used in place of real data to train a self-supervised discriminative model. Synthetic images shared between institutions are subject to fewer regulatory barriers than would be the case with real clinical images. Furthermore, we showcase the ability to precisely control image generation using a small set of annotated images, which can be used for both data augmentation and educational purposes. Testing on a cell segmentation task, a mask-guided PixCell enables targeted data augmentation, improving downstream performance. Finally, we demonstrate PixCell's ability to use H\&E structural staining to infer results from molecular marker studies; we use this capability to infer IHC staining from H\&E images. Our trained models are publicly released to accelerate research in computational pathology.
Improving Contrastive Learning for Referring Expression Counting
May 28, 2025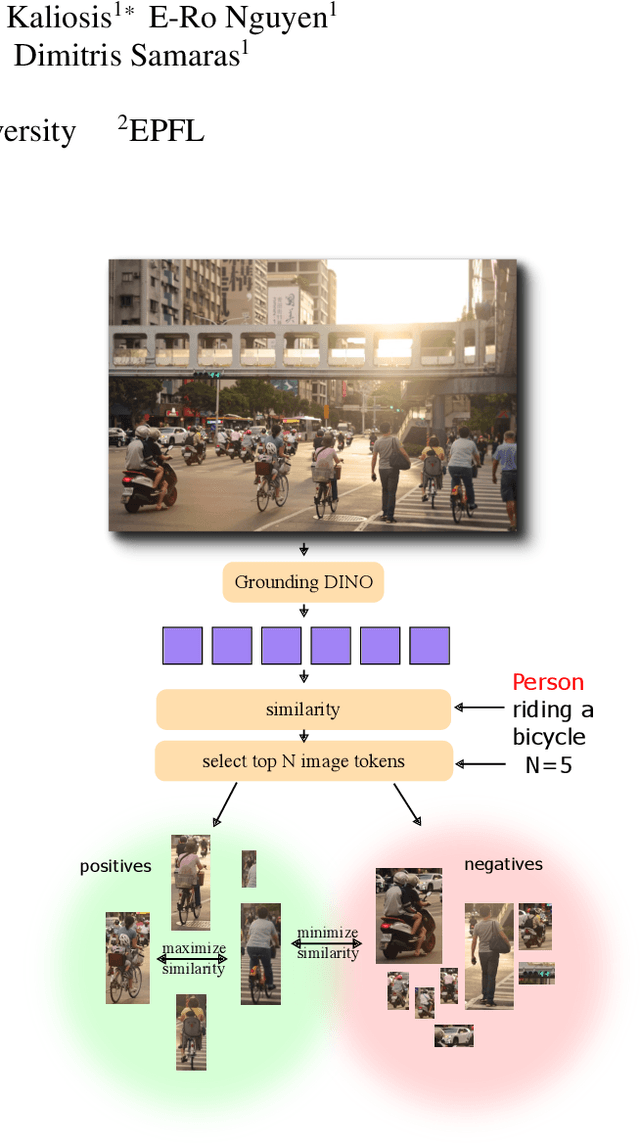
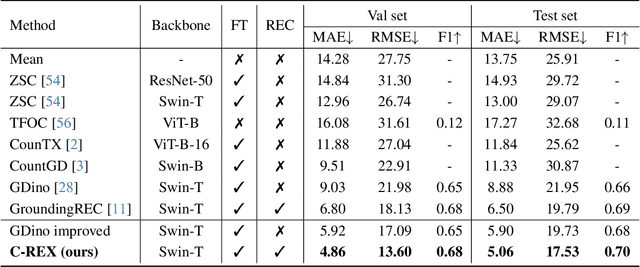

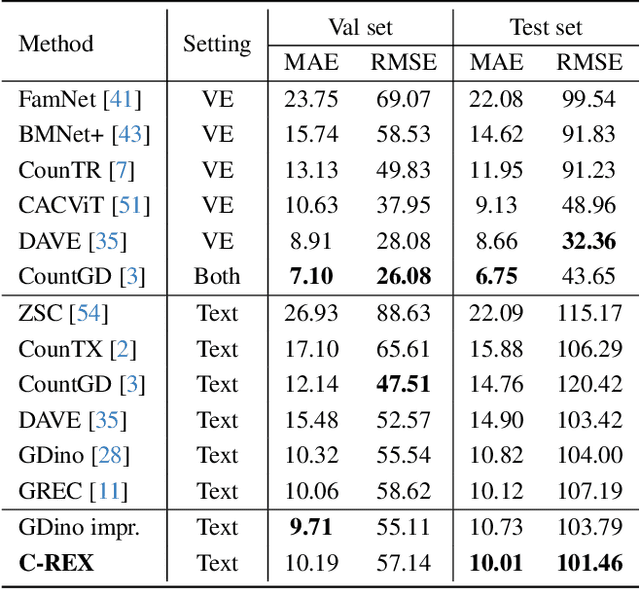
Abstract:Object counting has progressed from class-specific models, which count only known categories, to class-agnostic models that generalize to unseen categories. The next challenge is Referring Expression Counting (REC), where the goal is to count objects based on fine-grained attributes and contextual differences. Existing methods struggle with distinguishing visually similar objects that belong to the same category but correspond to different referring expressions. To address this, we propose C-REX, a novel contrastive learning framework, based on supervised contrastive learning, designed to enhance discriminative representation learning. Unlike prior works, C-REX operates entirely within the image space, avoiding the misalignment issues of image-text contrastive learning, thus providing a more stable contrastive signal. It also guarantees a significantly larger pool of negative samples, leading to improved robustness in the learned representations. Moreover, we showcase that our framework is versatile and generic enough to be applied to other similar tasks like class-agnostic counting. To support our approach, we analyze the key components of sota detection-based models and identify that detecting object centroids instead of bounding boxes is the key common factor behind their success in counting tasks. We use this insight to design a simple yet effective detection-based baseline to build upon. Our experiments show that C-REX achieves state-of-the-art results in REC, outperforming previous methods by more than 22\% in MAE and more than 10\% in RMSE, while also demonstrating strong performance in class-agnostic counting. Code is available at https://github.com/cvlab-stonybrook/c-rex.
GeoMaNO: Geometric Mamba Neural Operator for Partial Differential Equations
May 17, 2025Abstract:The neural operator (NO) framework has emerged as a powerful tool for solving partial differential equations (PDEs). Recent NOs are dominated by the Transformer architecture, which offers NOs the capability to capture long-range dependencies in PDE dynamics. However, existing Transformer-based NOs suffer from quadratic complexity, lack geometric rigor, and thus suffer from sub-optimal performance on regular grids. As a remedy, we propose the Geometric Mamba Neural Operator (GeoMaNO) framework, which empowers NOs with Mamba's modeling capability, linear complexity, plus geometric rigor. We evaluate GeoMaNO's performance on multiple standard and popularly employed PDE benchmarks, spanning from Darcy flow problems to Navier-Stokes problems. GeoMaNO improves existing baselines in solution operator approximation by as much as 58.9%.
PathSegDiff: Pathology Segmentation using Diffusion model representations
Apr 09, 2025Abstract:Image segmentation is crucial in many computational pathology pipelines, including accurate disease diagnosis, subtyping, outcome, and survivability prediction. The common approach for training a segmentation model relies on a pre-trained feature extractor and a dataset of paired image and mask annotations. These are used to train a lightweight prediction model that translates features into per-pixel classes. The choice of the feature extractor is central to the performance of the final segmentation model, and recent literature has focused on finding tasks to pre-train the feature extractor. In this paper, we propose PathSegDiff, a novel approach for histopathology image segmentation that leverages Latent Diffusion Models (LDMs) as pre-trained featured extractors. Our method utilizes a pathology-specific LDM, guided by a self-supervised encoder, to extract rich semantic information from H\&E stained histopathology images. We employ a simple, fully convolutional network to process the features extracted from the LDM and generate segmentation masks. Our experiments demonstrate significant improvements over traditional methods on the BCSS and GlaS datasets, highlighting the effectiveness of domain-specific diffusion pre-training in capturing intricate tissue structures and enhancing segmentation accuracy in histopathology images.
Few-shot Personalized Scanpath Prediction
Apr 07, 2025Abstract:A personalized model for scanpath prediction provides insights into the visual preferences and attention patterns of individual subjects. However, existing methods for training scanpath prediction models are data-intensive and cannot be effectively personalized to new individuals with only a few available examples. In this paper, we propose few-shot personalized scanpath prediction task (FS-PSP) and a novel method to address it, which aims to predict scanpaths for an unseen subject using minimal support data of that subject's scanpath behavior. The key to our method's adaptability is the Subject-Embedding Network (SE-Net), specifically designed to capture unique, individualized representations for each subject's scanpaths. SE-Net generates subject embeddings that effectively distinguish between subjects while minimizing variability among scanpaths from the same individual. The personalized scanpath prediction model is then conditioned on these subject embeddings to produce accurate, personalized results. Experiments on multiple eye-tracking datasets demonstrate that our method excels in FS-PSP settings and does not require any fine-tuning steps at test time. Code is available at: https://github.com/cvlab-stonybrook/few-shot-scanpath
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge