Meritxell Bach Cuadra
CIBM Center for Biomedical Imaging, Switzerland, Department of Radiology, Lausanne University Hospital
From 100,000+ images to winning the first brain MRI foundation model challenges: Sharing lessons and models
Jan 19, 2026Abstract:Developing Foundation Models for medical image analysis is essential to overcome the unique challenges of radiological tasks. The first challenges of this kind for 3D brain MRI, SSL3D and FOMO25, were held at MICCAI 2025. Our solution ranked first in tracks of both contests. It relies on a U-Net CNN architecture combined with strategies leveraging anatomical priors and neuroimaging domain knowledge. Notably, our models trained 1-2 orders of magnitude faster and were 10 times smaller than competing transformer-based approaches. Models are available here: https://github.com/jbanusco/BrainFM4Challenges.
Fetpype: An Open-Source Pipeline for Reproducible Fetal Brain MRI Analysis
Dec 19, 2025Abstract:Fetal brain Magnetic Resonance Imaging (MRI) is crucial for assessing neurodevelopment in utero. However, analyzing this data presents significant challenges due to fetal motion, low signal-to-noise ratio, and the need for complex multi-step processing, including motion correction, super-resolution reconstruction, segmentation, and surface extraction. While various specialized tools exist for individual steps, integrating them into robust, reproducible, and user-friendly workflows that go from raw images to processed volumes is not straightforward. This lack of standardization hinders reproducibility across studies and limits the adoption of advanced analysis techniques for researchers and clinicians. To address these challenges, we introduce Fetpype, an open-source Python library designed to streamline and standardize the preprocessing and analysis of T2-weighted fetal brain MRI data. Fetpype is publicly available on GitHub at https://github.com/fetpype/fetpype.
Causal Attribution of Model Performance Gaps in Medical Imaging Under Distribution Shifts
Dec 09, 2025Abstract:Deep learning models for medical image segmentation suffer significant performance drops due to distribution shifts, but the causal mechanisms behind these drops remain poorly understood. We extend causal attribution frameworks to high-dimensional segmentation tasks, quantifying how acquisition protocols and annotation variability independently contribute to performance degradation. We model the data-generating process through a causal graph and employ Shapley values to fairly attribute performance changes to individual mechanisms. Our framework addresses unique challenges in medical imaging: high-dimensional outputs, limited samples, and complex mechanism interactions. Validation on multiple sclerosis (MS) lesion segmentation across 4 centers and 7 annotators reveals context-dependent failure modes: annotation protocol shifts dominate when crossing annotators (7.4% $\pm$ 8.9% DSC attribution), while acquisition shifts dominate when crossing imaging centers (6.5% $\pm$ 9.1%). This mechanism-specific quantification enables practitioners to prioritize targeted interventions based on deployment context.
Benchmarking and Explaining Deep Learning Cortical Lesion MRI Segmentation in Multiple Sclerosis
Jul 16, 2025Abstract:Cortical lesions (CLs) have emerged as valuable biomarkers in multiple sclerosis (MS), offering high diagnostic specificity and prognostic relevance. However, their routine clinical integration remains limited due to subtle magnetic resonance imaging (MRI) appearance, challenges in expert annotation, and a lack of standardized automated methods. We propose a comprehensive multi-centric benchmark of CL detection and segmentation in MRI. A total of 656 MRI scans, including clinical trial and research data from four institutions, were acquired at 3T and 7T using MP2RAGE and MPRAGE sequences with expert-consensus annotations. We rely on the self-configuring nnU-Net framework, designed for medical imaging segmentation, and propose adaptations tailored to the improved CL detection. We evaluated model generalization through out-of-distribution testing, demonstrating strong lesion detection capabilities with an F1-score of 0.64 and 0.5 in and out of the domain, respectively. We also analyze internal model features and model errors for a better understanding of AI decision-making. Our study examines how data variability, lesion ambiguity, and protocol differences impact model performance, offering future recommendations to address these barriers to clinical adoption. To reinforce the reproducibility, the implementation and models will be publicly accessible and ready to use at https://github.com/Medical-Image-Analysis-Laboratory/ and https://doi.org/10.5281/zenodo.15911797.
ConfLUNet: Multiple sclerosis lesion instance segmentation in presence of confluent lesions
May 28, 2025Abstract:Accurate lesion-level segmentation on MRI is critical for multiple sclerosis (MS) diagnosis, prognosis, and disease monitoring. However, current evaluation practices largely rely on semantic segmentation post-processed with connected components (CC), which cannot separate confluent lesions (aggregates of confluent lesion units, CLUs) due to reliance on spatial connectivity. To address this misalignment with clinical needs, we introduce formal definitions of CLUs and associated CLU-aware detection metrics, and include them in an exhaustive instance segmentation evaluation framework. Within this framework, we systematically evaluate CC and post-processing-based Automated Confluent Splitting (ACLS), the only existing methods for lesion instance segmentation in MS. Our analysis reveals that CC consistently underestimates CLU counts, while ACLS tends to oversplit lesions, leading to overestimated lesion counts and reduced precision. To overcome these limitations, we propose ConfLUNet, the first end-to-end instance segmentation framework for MS lesions. ConfLUNet jointly optimizes lesion detection and delineation from a single FLAIR image. Trained on 50 patients, ConfLUNet significantly outperforms CC and ACLS on the held-out test set (n=13) in instance segmentation (Panoptic Quality: 42.0% vs. 37.5%/36.8%; p = 0.017/0.005) and lesion detection (F1: 67.3% vs. 61.6%/59.9%; p = 0.028/0.013). For CLU detection, ConfLUNet achieves the highest F1[CLU] (81.5%), improving recall over CC (+12.5%, p = 0.015) and precision over ACLS (+31.2%, p = 0.003). By combining rigorous definitions, new CLU-aware metrics, a reproducible evaluation framework, and the first dedicated end-to-end model, this work lays the foundation for lesion instance segmentation in MS.
Meta-learning Slice-to-Volume Reconstruction in Fetal Brain MRI using Implicit Neural Representations
May 14, 2025



Abstract:High-resolution slice-to-volume reconstruction (SVR) from multiple motion-corrupted low-resolution 2D slices constitutes a critical step in image-based diagnostics of moving subjects, such as fetal brain Magnetic Resonance Imaging (MRI). Existing solutions struggle with image artifacts and severe subject motion or require slice pre-alignment to achieve satisfying reconstruction performance. We propose a novel SVR method to enable fast and accurate MRI reconstruction even in cases of severe image and motion corruption. Our approach performs motion correction, outlier handling, and super-resolution reconstruction with all operations being entirely based on implicit neural representations. The model can be initialized with task-specific priors through fully self-supervised meta-learning on either simulated or real-world data. In extensive experiments including over 480 reconstructions of simulated and clinical MRI brain data from different centers, we prove the utility of our method in cases of severe subject motion and image artifacts. Our results demonstrate improvements in reconstruction quality, especially in the presence of severe motion, compared to state-of-the-art methods, and up to 50% reduction in reconstruction time.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


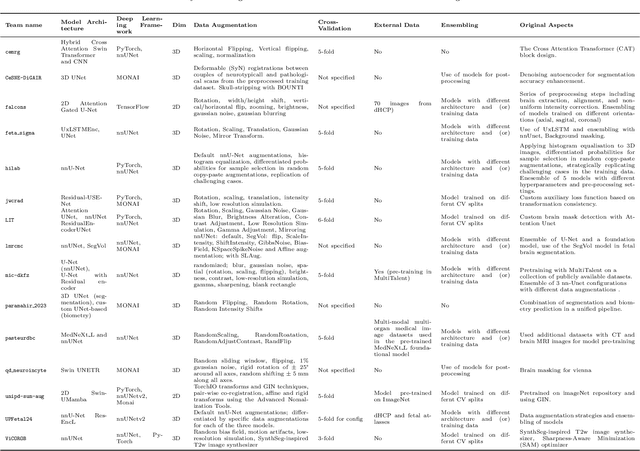
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Towards contrast- and pathology-agnostic clinical fetal brain MRI segmentation using SynthSeg
Apr 14, 2025



Abstract:Magnetic resonance imaging (MRI) has played a crucial role in fetal neurodevelopmental research. Structural annotations of MR images are an important step for quantitative analysis of the developing human brain, with Deep learning providing an automated alternative for this otherwise tedious manual process. However, segmentation performances of Convolutional Neural Networks often suffer from domain shift, where the network fails when applied to subjects that deviate from the distribution with which it is trained on. In this work, we aim to train networks capable of automatically segmenting fetal brain MRIs with a wide range of domain shifts pertaining to differences in subject physiology and acquisition environments, in particular shape-based differences commonly observed in pathological cases. We introduce a novel data-driven train-time sampling strategy that seeks to fully exploit the diversity of a given training dataset to enhance the domain generalizability of the trained networks. We adapted our sampler, together with other existing data augmentation techniques, to the SynthSeg framework, a generator that utilizes domain randomization to generate diverse training data, and ran thorough experimentations and ablation studies on a wide range of training/testing data to test the validity of the approaches. Our networks achieved notable improvements in the segmentation quality on testing subjects with intense anatomical abnormalities (p < 1e-4), though at the cost of a slighter decrease in performance in cases with fewer abnormalities. Our work also lays the foundation for future works on creating and adapting data-driven sampling strategies for other training pipelines.
Explainability of AI Uncertainty: Application to Multiple Sclerosis Lesion Segmentation on MRI
Apr 07, 2025



Abstract:Trustworthy artificial intelligence (AI) is essential in healthcare, particularly for high-stakes tasks like medical image segmentation. Explainable AI and uncertainty quantification significantly enhance AI reliability by addressing key attributes such as robustness, usability, and explainability. Despite extensive technical advances in uncertainty quantification for medical imaging, understanding the clinical informativeness and interpretability of uncertainty remains limited. This study introduces a novel framework to explain the potential sources of predictive uncertainty, specifically in cortical lesion segmentation in multiple sclerosis using deep ensembles. The proposed analysis shifts the focus from the uncertainty-error relationship towards relevant medical and engineering factors. Our findings reveal that instance-wise uncertainty is strongly related to lesion size, shape, and cortical involvement. Expert rater feedback confirms that similar factors impede annotator confidence. Evaluations conducted on two datasets (206 patients, almost 2000 lesions) under both in-domain and distribution-shift conditions highlight the utility of the framework in different scenarios.
Automatic quality control in multi-centric fetal brain MRI super-resolution reconstruction
Mar 13, 2025Abstract:Quality control (QC) has long been considered essential to guarantee the reliability of neuroimaging studies. It is particularly important for fetal brain MRI, where acquisitions and image processing techniques are less standardized than in adult imaging. In this work, we focus on automated quality control of super-resolution reconstruction (SRR) volumes of fetal brain MRI, an important processing step where multiple stacks of thick 2D slices are registered together and combined to build a single, isotropic and artifact-free T2 weighted volume. We propose FetMRQC$_{SR}$, a machine-learning method that extracts more than 100 image quality metrics to predict image quality scores using a random forest model. This approach is well suited to a problem that is high dimensional, with highly heterogeneous data and small datasets. We validate FetMRQC$_{SR}$ in an out-of-domain (OOD) setting and report high performance (ROC AUC = 0.89), even when faced with data from an unknown site or SRR method. We also investigate failure cases and show that they occur in $45\%$ of the images due to ambiguous configurations for which the rating from the expert is arguable. These results are encouraging and illustrate how a non deep learning-based method like FetMRQC$_{SR}$ is well suited to this multifaceted problem. Our tool, along with all the code used to generate, train and evaluate the model will be released upon acceptance of the paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge