Joseph V. Hajnal
Meta-learning Slice-to-Volume Reconstruction in Fetal Brain MRI using Implicit Neural Representations
May 14, 2025



Abstract:High-resolution slice-to-volume reconstruction (SVR) from multiple motion-corrupted low-resolution 2D slices constitutes a critical step in image-based diagnostics of moving subjects, such as fetal brain Magnetic Resonance Imaging (MRI). Existing solutions struggle with image artifacts and severe subject motion or require slice pre-alignment to achieve satisfying reconstruction performance. We propose a novel SVR method to enable fast and accurate MRI reconstruction even in cases of severe image and motion corruption. Our approach performs motion correction, outlier handling, and super-resolution reconstruction with all operations being entirely based on implicit neural representations. The model can be initialized with task-specific priors through fully self-supervised meta-learning on either simulated or real-world data. In extensive experiments including over 480 reconstructions of simulated and clinical MRI brain data from different centers, we prove the utility of our method in cases of severe subject motion and image artifacts. Our results demonstrate improvements in reconstruction quality, especially in the presence of severe motion, compared to state-of-the-art methods, and up to 50% reduction in reconstruction time.
L-FUSION: Laplacian Fetal Ultrasound Segmentation & Uncertainty Estimation
Mar 07, 2025

Abstract:Accurate analysis of prenatal ultrasound (US) is essential for early detection of developmental anomalies. However, operator dependency and technical limitations (e.g. intrinsic artefacts and effects, setting errors) can complicate image interpretation and the assessment of diagnostic uncertainty. We present L-FUSION (Laplacian Fetal US Segmentation with Integrated FoundatiON models), a framework that integrates uncertainty quantification through unsupervised, normative learning and large-scale foundation models for robust segmentation of fetal structures in normal and pathological scans. We propose to utilise the aleatoric logit distributions of Stochastic Segmentation Networks and Laplace approximations with fast Hessian estimations to estimate epistemic uncertainty only from the segmentation head. This enables us to achieve reliable abnormality quantification for instant diagnostic feedback. Combined with an integrated Dropout component, L-FUSION enables reliable differentiation of lesions from normal fetal anatomy with enhanced uncertainty maps and segmentation counterfactuals in US imaging. It improves epistemic and aleatoric uncertainty interpretation and removes the need for manual disease-labelling. Evaluations across multiple datasets show that L-FUSION achieves superior segmentation accuracy and consistent uncertainty quantification, supporting on-site decision-making and offering a scalable solution for advancing fetal ultrasound analysis in clinical settings.
DCRA-Net: Attention-Enabled Reconstruction Model for Dynamic Fetal Cardiac MRI
Dec 19, 2024



Abstract:Dynamic fetal heart magnetic resonance imaging (MRI) presents unique challenges due to the fast heart rate of the fetus compared to adult subjects and uncontrolled fetal motion. This requires high temporal and spatial resolutions over a large field of view, in order to encompass surrounding maternal anatomy. In this work, we introduce Dynamic Cardiac Reconstruction Attention Network (DCRA-Net) - a novel deep learning model that employs attention mechanisms in spatial and temporal domains and temporal frequency representation of data to reconstruct the dynamics of the fetal heart from highly accelerated free-running (non-gated) MRI acquisitions. DCRA-Net was trained on retrospectively undersampled complex-valued cardiac MRIs from 42 fetal subjects and separately from 153 adult subjects, and evaluated on data from 14 fetal and 39 adult subjects respectively. Its performance was compared to L+S and k-GIN methods in both fetal and adult cases for an undersampling factor of 8x. The proposed network performed better than the comparators for both fetal and adult data, for both regular lattice and centrally weighted random undersampling. Aliased signals due to the undersampling were comprehensively resolved, and both the spatial details of the heart and its temporal dynamics were recovered with high fidelity. The highest performance was achieved when using lattice undersampling, data consistency and temporal frequency representation, yielding PSNR of 38 for fetal and 35 for adult cases. Our method is publicly available at https://github.com/denproc/DCRA-Net.
CINA: Conditional Implicit Neural Atlas for Spatio-Temporal Representation of Fetal Brains
Mar 13, 2024



Abstract:We introduce a conditional implicit neural atlas (CINA) for spatio-temporal atlas generation from Magnetic Resonance Images (MRI) of the neurotypical and pathological fetal brain, that is fully independent of affine or non-rigid registration. During training, CINA learns a general representation of the fetal brain and encodes subject specific information into latent code. After training, CINA can construct a faithful atlas with tissue probability maps of the fetal brain for any gestational age (GA) and anatomical variation covered within the training domain. Thus, CINA is competent to represent both, neurotypical and pathological brains. Furthermore, a trained CINA model can be fit to brain MRI of unseen subjects via test-time optimization of the latent code. CINA can then produce probabilistic tissue maps tailored to a particular subject. We evaluate our method on a total of 198 T2 weighted MRI of normal and abnormal fetal brains from the dHCP and FeTA datasets. We demonstrate CINA's capability to represent a fetal brain atlas that can be flexibly conditioned on GA and on anatomical variations like ventricular volume or degree of cortical folding, making it a suitable tool for modeling both neurotypical and pathological brains. We quantify the fidelity of our atlas by means of tissue segmentation and age prediction and compare it to an established baseline. CINA demonstrates superior accuracy for neurotypical brains and pathological brains with ventriculomegaly. Moreover, CINA scores a mean absolute error of 0.23 weeks in fetal brain age prediction, further confirming an accurate representation of fetal brain development.
An automated pipeline for quantitative T2* fetal body MRI and segmentation at low field
Aug 09, 2023Abstract:Fetal Magnetic Resonance Imaging at low field strengths is emerging as an exciting direction in perinatal health. Clinical low field (0.55T) scanners are beneficial for fetal imaging due to their reduced susceptibility-induced artefacts, increased T2* values, and wider bore (widening access for the increasingly obese pregnant population). However, the lack of standard automated image processing tools such as segmentation and reconstruction hampers wider clinical use. In this study, we introduce a semi-automatic pipeline using quantitative MRI for the fetal body at low field strength resulting in fast and detailed quantitative T2* relaxometry analysis of all major fetal body organs. Multi-echo dynamic sequences of the fetal body were acquired and reconstructed into a single high-resolution volume using deformable slice-to-volume reconstruction, generating both structural and quantitative T2* 3D volumes. A neural network trained using a semi-supervised approach was created to automatically segment these fetal body 3D volumes into ten different organs (resulting in dice values > 0.74 for 8 out of 10 organs). The T2* values revealed a strong relationship with GA in the lungs, liver, and kidney parenchyma (R^2>0.5). This pipeline was used successfully for a wide range of GAs (17-40 weeks), and is robust to motion artefacts. Low field fetal MRI can be used to perform advanced MRI analysis, and is a viable option for clinical scanning.
Placenta Segmentation in Ultrasound Imaging: Addressing Sources of Uncertainty and Limited Field-of-View
Jun 29, 2022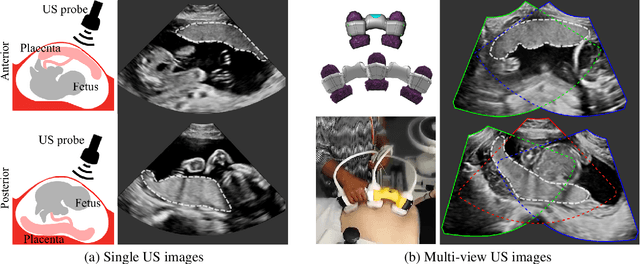
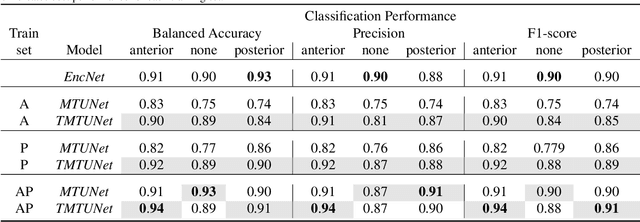
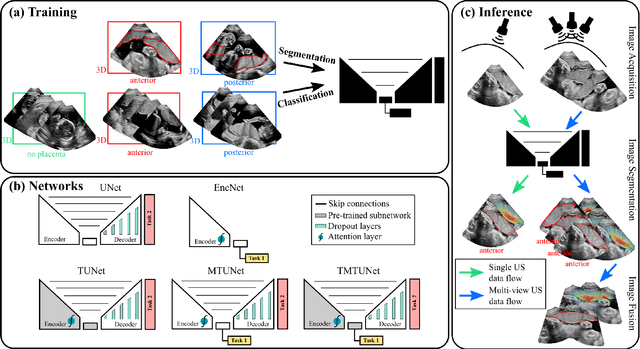
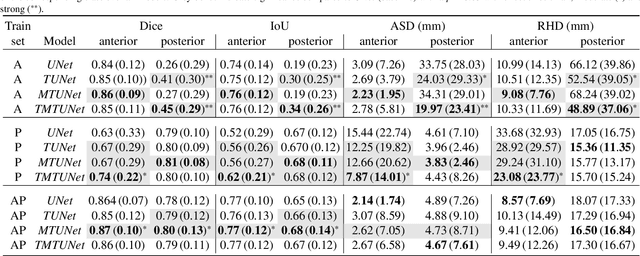
Abstract:Automatic segmentation of the placenta in fetal ultrasound (US) is challenging due to the (i) high diversity of placenta appearance, (ii) the restricted quality in US resulting in highly variable reference annotations, and (iii) the limited field-of-view of US prohibiting whole placenta assessment at late gestation. In this work, we address these three challenges with a multi-task learning approach that combines the classification of placental location (e.g., anterior, posterior) and semantic placenta segmentation in a single convolutional neural network. Through the classification task the model can learn from larger and more diverse datasets while improving the accuracy of the segmentation task in particular in limited training set conditions. With this approach we investigate the variability in annotations from multiple raters and show that our automatic segmentations (Dice of 0.86 for anterior and 0.83 for posterior placentas) achieve human-level performance as compared to intra- and inter-observer variability. Lastly, our approach can deliver whole placenta segmentation using a multi-view US acquisition pipeline consisting of three stages: multi-probe image acquisition, image fusion and image segmentation. This results in high quality segmentation of larger structures such as the placenta in US with reduced image artifacts which are beyond the field-of-view of single probes.
Fetal MRI by robust deep generative prior reconstruction and diffeomorphic registration: application to gestational age prediction
Oct 29, 2021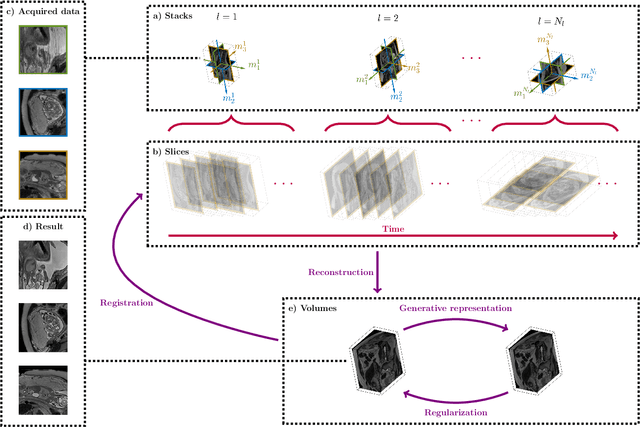
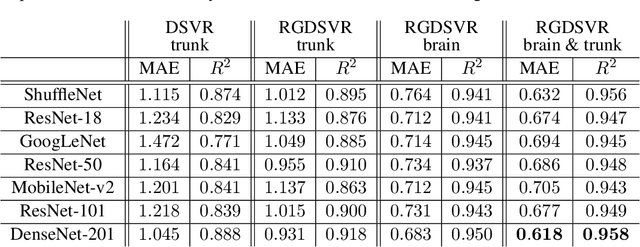

Abstract:Magnetic resonance imaging of whole fetal body and placenta is limited by different sources of motion affecting the womb. Usual scanning techniques employ single-shot multi-slice sequences where anatomical information in different slices may be subject to different deformations, contrast variations or artifacts. Volumetric reconstruction formulations have been proposed to correct for these factors, but they must accommodate a non-homogeneous and non-isotropic sampling, so regularization becomes necessary. Thus, in this paper we propose a deep generative prior for robust volumetric reconstructions integrated with a diffeomorphic volume to slice registration method. Experiments are performed to validate our contributions and compare with a state of the art method in a cohort of $72$ fetal datasets in the range of $20-36$ weeks gestational age. Results suggest improved image resolution and more accurate prediction of gestational age at scan when comparing to a state of the art reconstruction method. In addition, gestational age prediction results from our volumetric reconstructions compare favourably with existing brain-based approaches, with boosted accuracy when integrating information of organs other than the brain. Namely, a mean absolute error of $0.618$ weeks ($R^2=0.958$) is achieved when combining fetal brain and trunk information.
Magnetization Transfer-Mediated MR Fingerprinting
Apr 06, 2021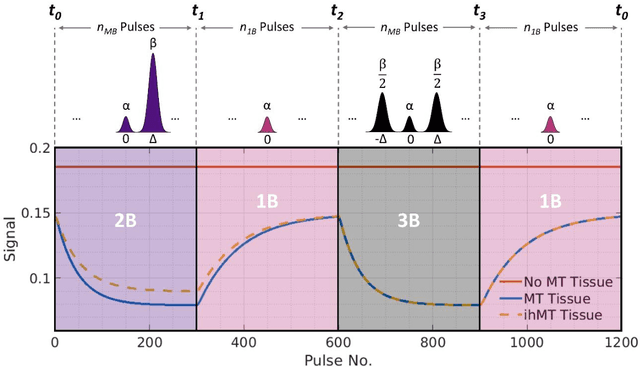
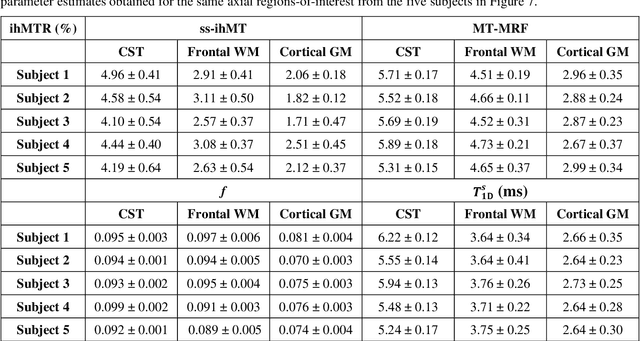
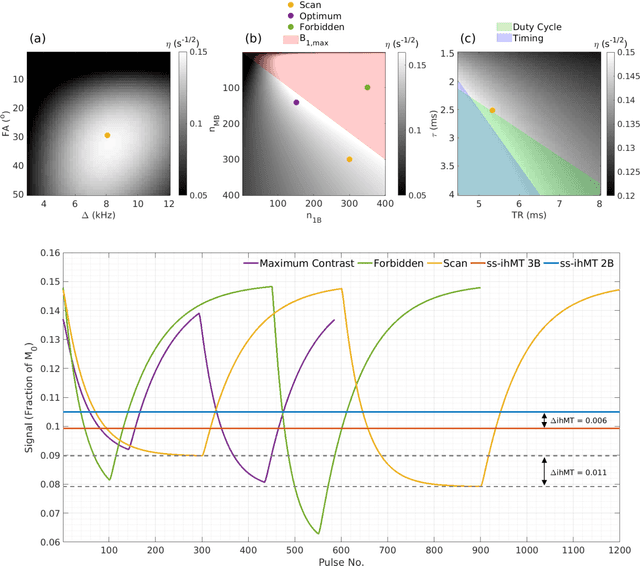
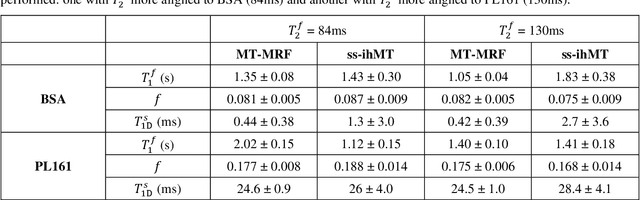
Abstract:Purpose: Magnetization transfer (MT) and inhomogeneous MT (ihMT) contrasts are used in MRI to provide information about macromolecular tissue content. In particular, MT is sensitive to macromolecules and ihMT appears to be specific to myelinated tissue. This study proposes a technique to characterize MT and ihMT properties from a single acquisition, producing both semiquantitative contrast ratios, and quantitative parameter maps. Theory and Methods: Building upon previous work that uses multiband radiofrequency (RF) pulses to efficiently generate ihMT contrast, we propose a cyclic-steady-state approach that cycles between multiband and single-band pulses to boost the achieved contrast. Resultant time-variable signals are reminiscent of a magnetic resonance fingerprinting (MRF) acquisition, except that the signal fluctuations are entirely mediated by magnetization transfer effects. A dictionary-based low-rank inversion method is used to reconstruct the resulting images and to produce both semiquantitative MT ratio (MTR) and ihMT ratio (ihMTR) maps, as well as quantitative parameter estimates corresponding to an ihMT tissue model. Results: Phantom and in vivo brain data acquired at 1.5T demonstrate the expected contrast trends, with ihMTR maps showing contrast more specific to white matter (WM), as has been reported by others. Quantitative estimation of semisolid fraction and dipolar T1 was also possible and yielded measurements consistent with literature values in the brain. Conclusions: By cycling between multiband and single-band pulses, an entirely magnetization transfer mediated 'fingerprinting' method was demonstrated. This proof-of-concept approach can be used to generate semiquantitative maps and quantitatively estimate some macromolecular specific tissue parameters.
Complementary Time-Frequency Domain Networks for Dynamic Parallel MR Image Reconstruction
Dec 22, 2020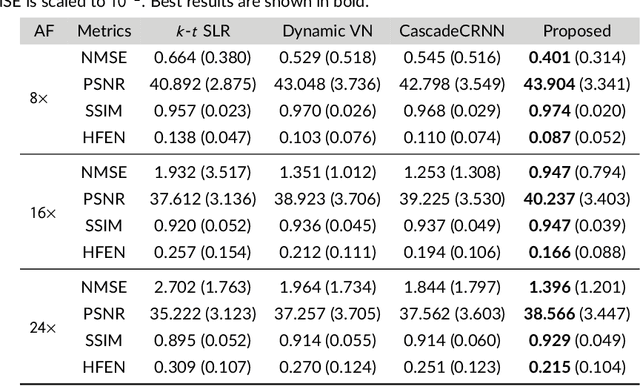
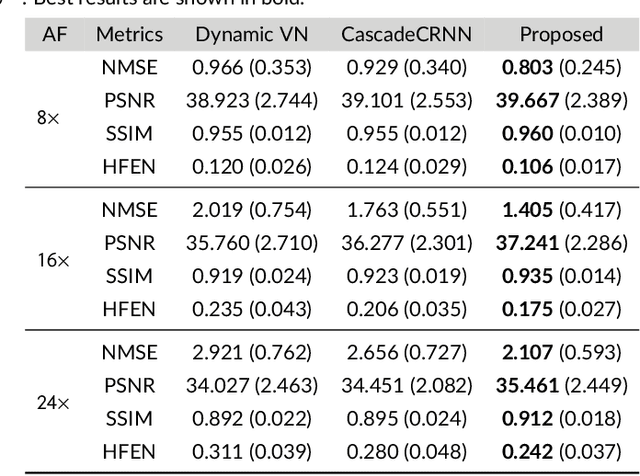
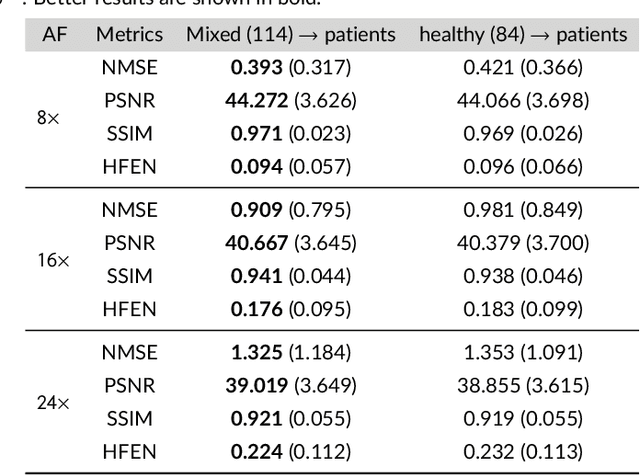
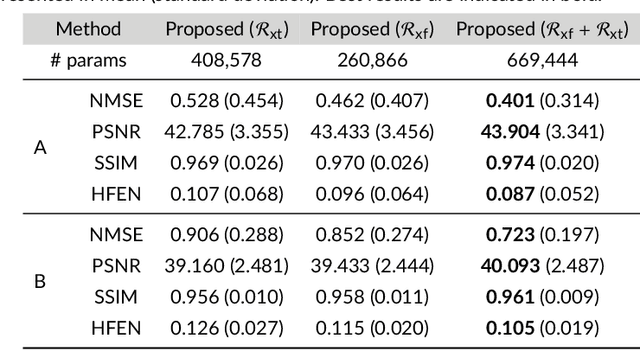
Abstract:Purpose: To introduce a novel deep learning based approach for fast and high-quality dynamic multi-coil MR reconstruction by learning a complementary time-frequency domain network that exploits spatio-temporal correlations simultaneously from complementary domains. Theory and Methods: Dynamic parallel MR image reconstruction is formulated as a multi-variable minimisation problem, where the data is regularised in combined temporal Fourier and spatial (x-f) domain as well as in spatio-temporal image (x-t) domain. An iterative algorithm based on variable splitting technique is derived, which alternates among signal de-aliasing steps in x-f and x-t spaces, a closed-form point-wise data consistency step and a weighted coupling step. The iterative model is embedded into a deep recurrent neural network which learns to recover the image via exploiting spatio-temporal redundancies in complementary domains. Results: Experiments were performed on two datasets of highly undersampled multi-coil short-axis cardiac cine MRI scans. Results demonstrate that our proposed method outperforms the current state-of-the-art approaches both quantitatively and qualitatively. The proposed model can also generalise well to data acquired from a different scanner and data with pathologies that were not seen in the training set. Conclusion: The work shows the benefit of reconstructing dynamic parallel MRI in complementary time-frequency domains with deep neural networks. The method can effectively and robustly reconstruct high-quality images from highly undersampled dynamic multi-coil data ($16 \times$ and $24 \times$ yielding 15s and 10s scan times respectively) with fast reconstruction speed (2.8s). This could potentially facilitate achieving fast single-breath-hold clinical 2D cardiac cine imaging.
Data consistency networks for (calibration-less) accelerated parallel MR image reconstruction
Sep 25, 2019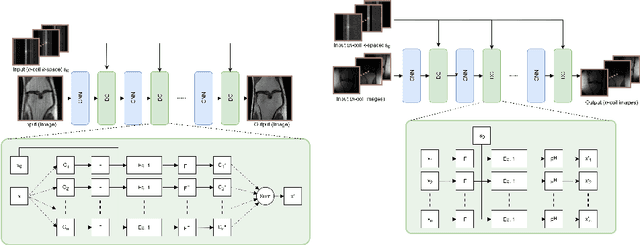
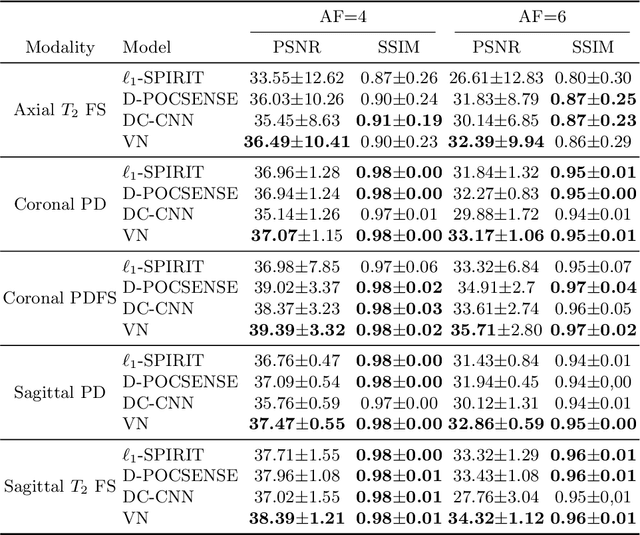
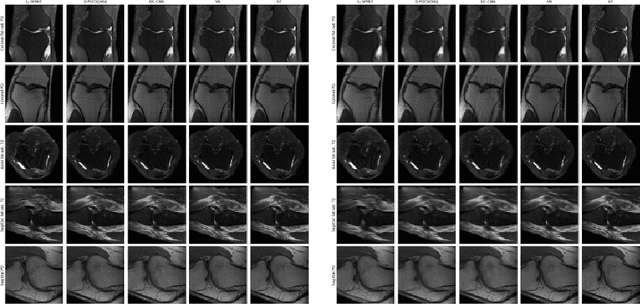
Abstract:We present simple reconstruction networks for multi-coil data by extending deep cascade of CNN's and exploiting the data consistency layer. In particular, we propose two variants, where one is inspired by POCSENSE and the other is calibration-less. We show that the proposed approaches are competitive relative to the state of the art both quantitatively and qualitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge