Improving Domain-Invariance in Self-Supervised Learning via Batch Styles Standardization
Mar 13, 2023Marin Scalbert, Maria Vakalopoulou, Florent Couzinié-Devy
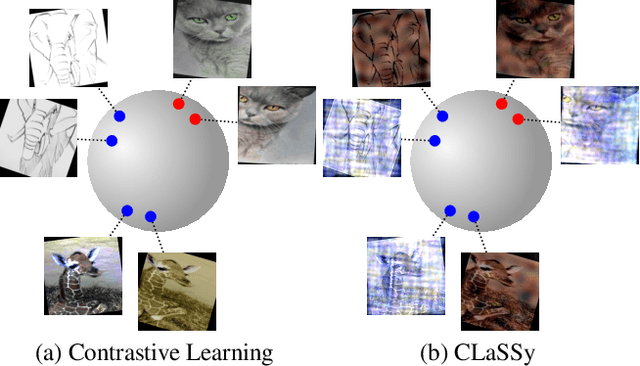
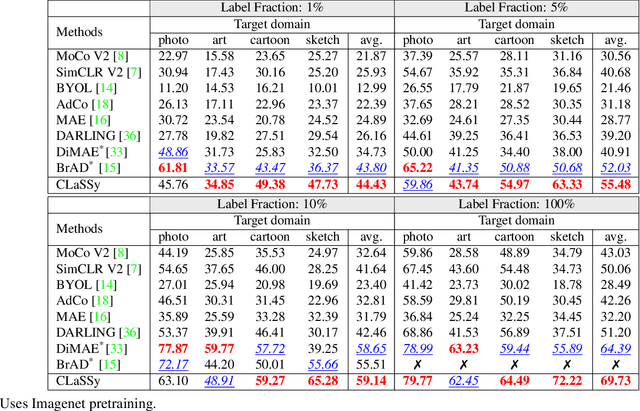
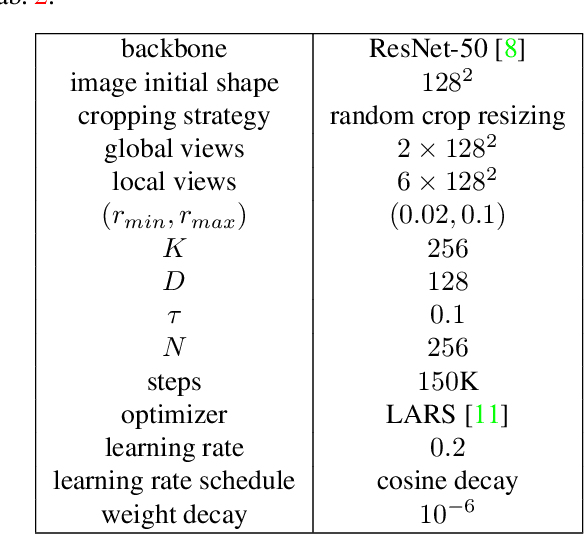
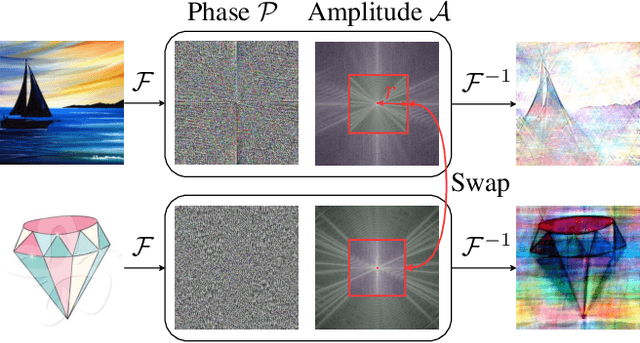
The recent rise of Self-Supervised Learning (SSL) as one of the preferred strategies for learning with limited labeled data, and abundant unlabeled data has led to the widespread use of these models. They are usually pretrained, finetuned, and evaluated on the same data distribution, i.e., within an in-distribution setting. However, they tend to perform poorly in out-of-distribution evaluation scenarios, a challenge that Unsupervised Domain Generalization (UDG) seeks to address. This paper introduces a novel method to standardize the styles of images in a batch. Batch styles standardization, relying on Fourier-based augmentations, promotes domain invariance in SSL by preventing spurious correlations from leaking into the features. The combination of batch styles standardization with the well-known contrastive-based method SimCLR leads to a novel UDG method named CLaSSy ($\textbf{C}$ontrastive $\textbf{L}$e$\textbf{a}$rning with $\textbf{S}$tandardized $\textbf{S}$t$\textbf{y}$les). CLaSSy offers serious advantages over prior methods, as it does not rely on domain labels and is scalable to handle a large number of domains. Experimental results on various UDG datasets demonstrate the superior performance of CLaSSy compared to existing UDG methods. Finally, the versatility of the proposed batch styles standardization is demonstrated by extending respectively the contrastive-based and non-contrastive-based SSL methods, SWaV and MSN, while considering different backbone architectures (convolutional-based, transformers-based).
Precise Location Matching Improves Dense Contrastive Learning in Digital Pathology
Dec 23, 2022Jingwei Zhang, Saarthak Kapse, Ke Ma, Prateek Prasanna, Maria Vakalopoulou, Joel Saltz, Dimitris Samaras
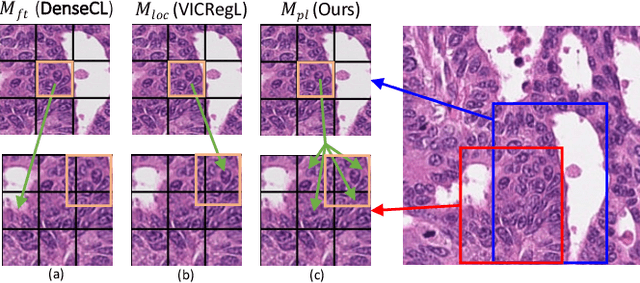
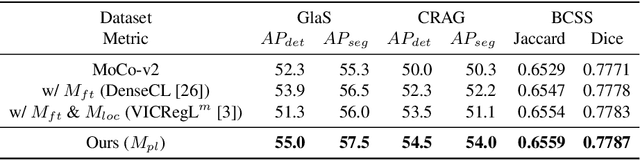
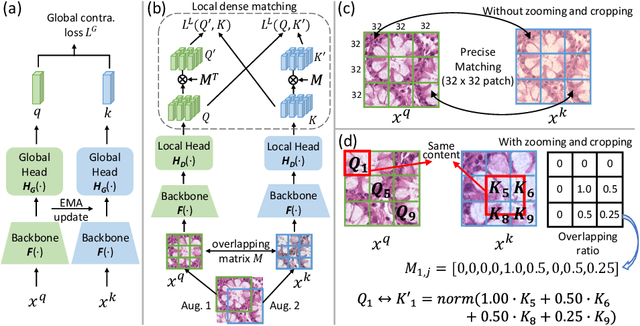

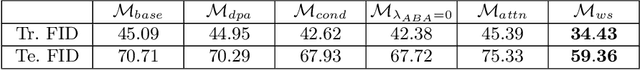
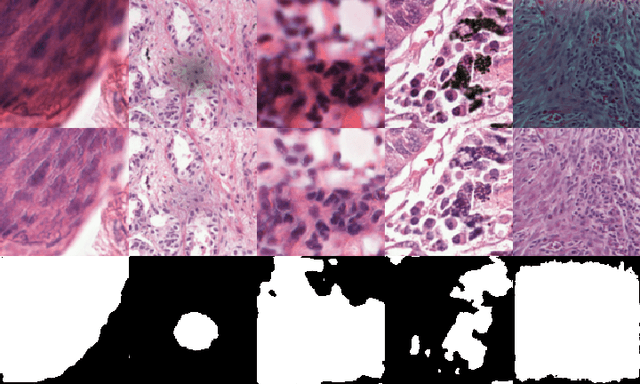
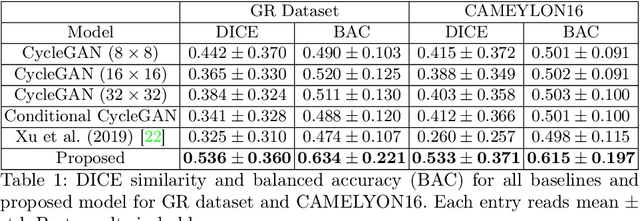
Dense prediction tasks such as segmentation and detection of pathological entities hold crucial clinical value in the digital pathology workflow. However, obtaining dense annotations on large cohorts is usually tedious and expensive. Contrastive learning (CL) is thus often employed to leverage large volumes of unlabeled data to pre-train the backbone network. To boost CL for dense prediction, some studies have proposed variations of dense matching objectives in pre-training. However, our analysis shows that employing existing dense matching strategies on histopathology images enforces invariance among incorrect pairs of dense features and, thus, is imprecise. To address this, we propose a precise location-based matching mechanism that utilizes the overlapping information between geometric transformations to precisely match regions in two augmentations. Extensive experiments on two pretraining datasets (TCGA-BRCA, NCT-CRC-HE) and three downstream datasets (GlaS, CRAG, BCSS) highlight the superiority of our method in semantic and instance segmentation tasks. Our method outperforms previous dense matching methods by up to 7.2 % in average precision for detection and 5.6 % in average precision for instance segmentation tasks. Additionally, by using our matching mechanism in the three popular contrastive learning frameworks, MoCo-v2, VICRegL and ConCL, the average precision in detection is improved by 0.7 % to 5.2 % and the average precision in segmentation is improved by 0.7 % to 4.0 %, demonstrating its generalizability.
Artifact Removal in Histopathology Images
Dec 16, 2022Cameron Dahan, Stergios Christodoulidis, Maria Vakalopoulou, Joseph Boyd
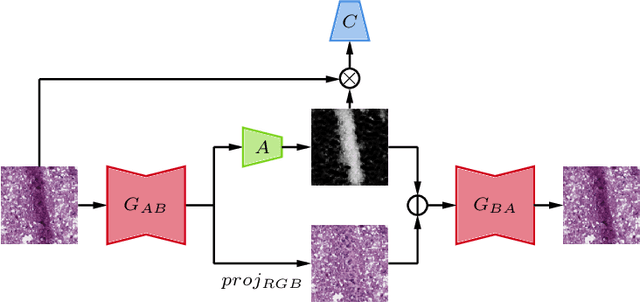
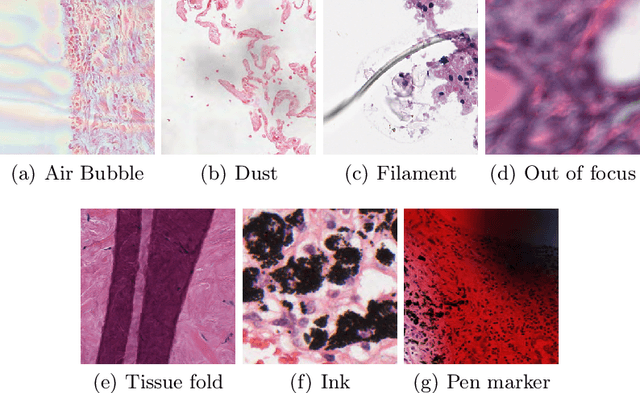
In the clinical setting of histopathology, whole-slide image (WSI) artifacts frequently arise, distorting regions of interest, and having a pernicious impact on WSI analysis. Image-to-image translation networks such as CycleGANs are in principle capable of learning an artifact removal function from unpaired data. However, we identify a surjection problem with artifact removal, and propose an weakly-supervised extension to CycleGAN to address this. We assemble a pan-cancer dataset comprising artifact and clean tiles from the TCGA database. Promising results highlight the soundness of our method.
Multi-center anatomical segmentation with heterogeneous labels via landmark-based models
Nov 14, 2022Nicolás Gaggion, Maria Vakalopoulou, Diego H. Milone, Enzo Ferrante
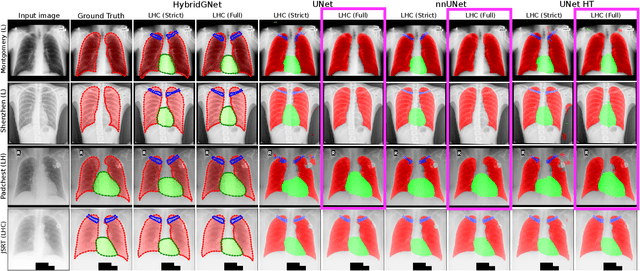
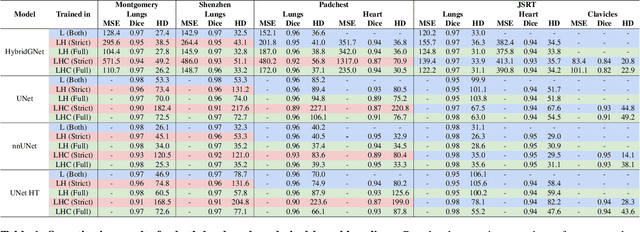
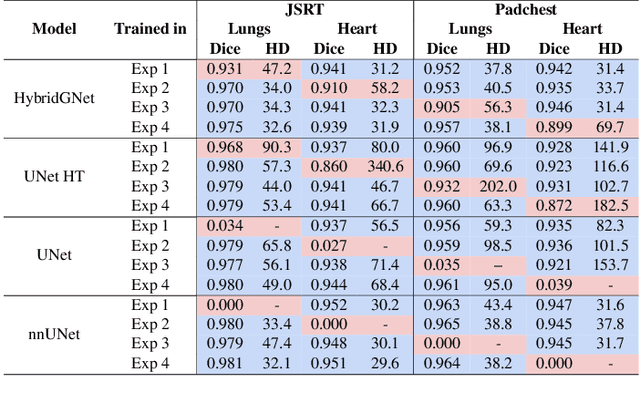
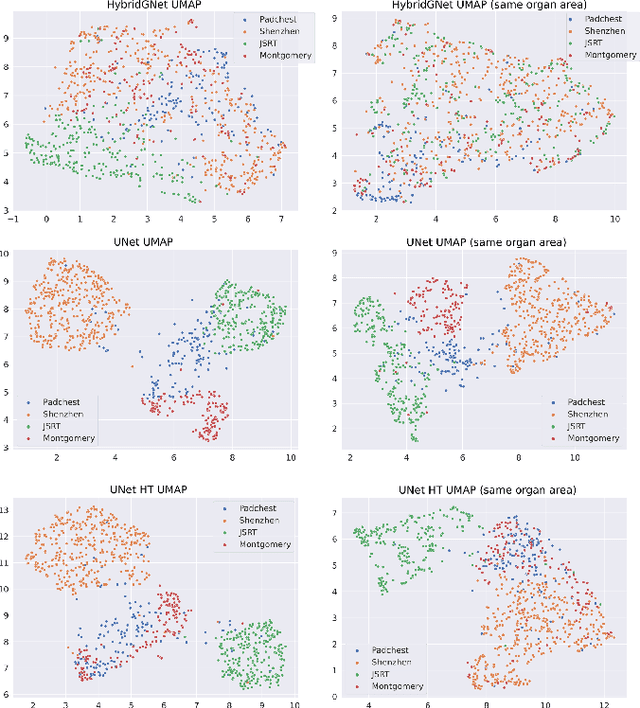
Learning anatomical segmentation from heterogeneous labels in multi-center datasets is a common situation encountered in clinical scenarios, where certain anatomical structures are only annotated in images coming from particular medical centers, but not in the full database. Here we first show how state-of-the-art pixel-level segmentation models fail in naively learning this task due to domain memorization issues and conflicting labels. We then propose to adopt HybridGNet, a landmark-based segmentation model which learns the available anatomical structures using graph-based representations. By analyzing the latent space learned by both models, we show that HybridGNet naturally learns more domain-invariant feature representations, and provide empirical evidence in the context of chest X-ray multiclass segmentation. We hope these insights will shed light on the training of deep learning models with heterogeneous labels from public and multi-center datasets.
Region-guided CycleGANs for Stain Transfer in Whole Slide Images
Aug 26, 2022Joseph Boyd, Irène Villa, Marie-Christine Mathieu, Eric Deutsch, Nikos Paragios, Maria Vakalopoulou, Stergios Christodoulidis
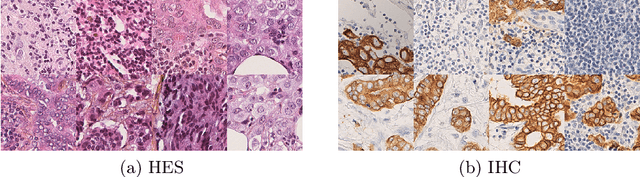
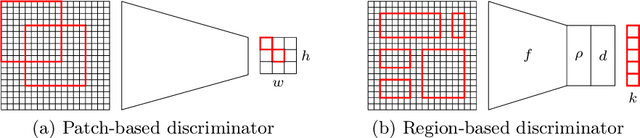
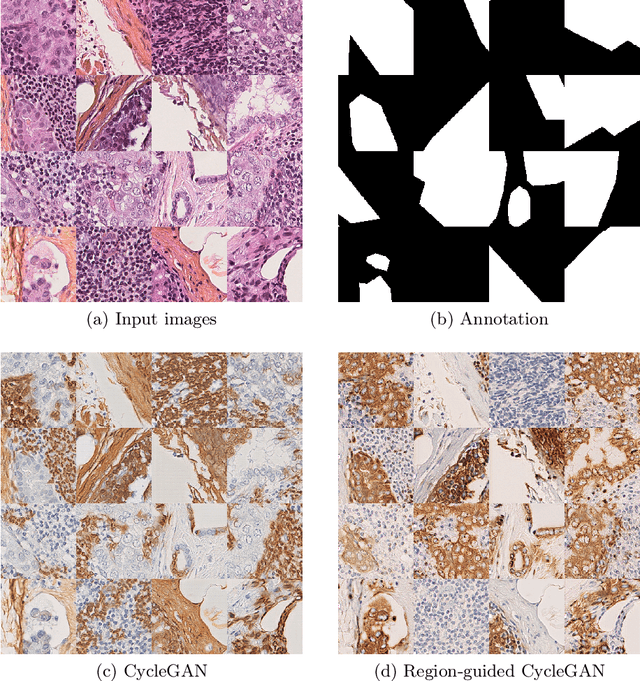
In whole slide imaging, commonly used staining techniques based on hematoxylin and eosin (H&E) and immunohistochemistry (IHC) stains accentuate different aspects of the tissue landscape. In the case of detecting metastases, IHC provides a distinct readout that is readily interpretable by pathologists. IHC, however, is a more expensive approach and not available at all medical centers. Virtually generating IHC images from H&E using deep neural networks thus becomes an attractive alternative. Deep generative models such as CycleGANs learn a semantically-consistent mapping between two image domains, while emulating the textural properties of each domain. They are therefore a suitable choice for stain transfer applications. However, they remain fully unsupervised, and possess no mechanism for enforcing biological consistency in stain transfer. In this paper, we propose an extension to CycleGANs in the form of a region of interest discriminator. This allows the CycleGAN to learn from unpaired datasets where, in addition, there is a partial annotation of objects for which one wishes to enforce consistency. We present a use case on whole slide images, where an IHC stain provides an experimentally generated signal for metastatic cells. We demonstrate the superiority of our approach over prior art in stain transfer on histopathology tiles over two datasets. Our code and model are available at https://github.com/jcboyd/miccai2022-roigan.
Gigapixel Whole-Slide Images Classification using Locally Supervised Learning
Jul 17, 2022Jingwei Zhang, Xin Zhang, Ke Ma, Rajarsi Gupta, Joel Saltz, Maria Vakalopoulou, Dimitris Samaras
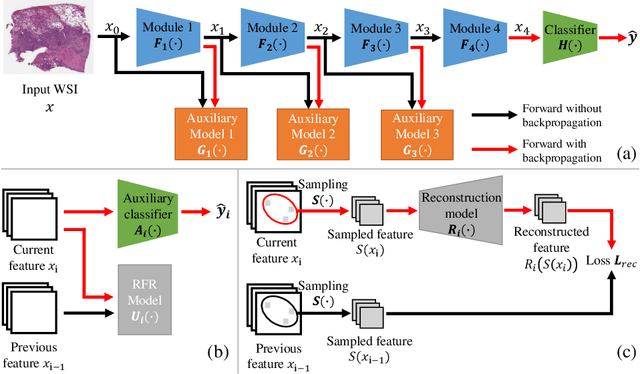
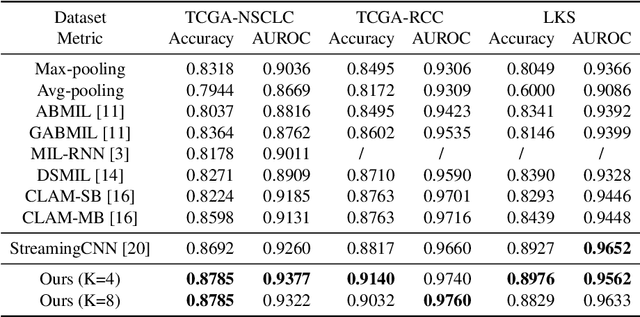

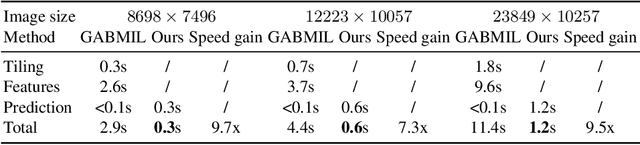
Histopathology whole slide images (WSIs) play a very important role in clinical studies and serve as the gold standard for many cancer diagnoses. However, generating automatic tools for processing WSIs is challenging due to their enormous sizes. Currently, to deal with this issue, conventional methods rely on a multiple instance learning (MIL) strategy to process a WSI at patch level. Although effective, such methods are computationally expensive, because tiling a WSI into patches takes time and does not explore the spatial relations between these tiles. To tackle these limitations, we propose a locally supervised learning framework which processes the entire slide by exploring the entire local and global information that it contains. This framework divides a pre-trained network into several modules and optimizes each module locally using an auxiliary model. We also introduce a random feature reconstruction unit (RFR) to preserve distinguishing features during training and improve the performance of our method by 1% to 3%. Extensive experiments on three publicly available WSI datasets: TCGA-NSCLC, TCGA-RCC and LKS, highlight the superiority of our method on different classification tasks. Our method outperforms the state-of-the-art MIL methods by 2% to 5% in accuracy, while being 7 to 10 times faster. Additionally, when dividing it into eight modules, our method requires as little as 20% of the total gpu memory required by end-to-end training. Our code is available at https://github.com/cvlab-stonybrook/local_learning_wsi.
Test-time image-to-image translation ensembling improves out-of-distribution generalization in histopathology
Jun 30, 2022Marin Scalbert, Maria Vakalopoulou, Florent Couzinié-Devy
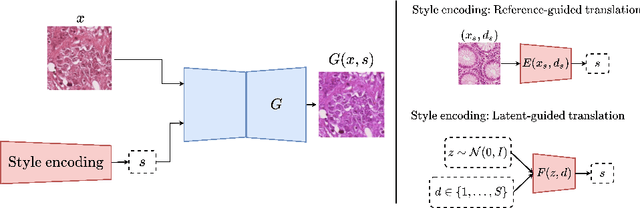
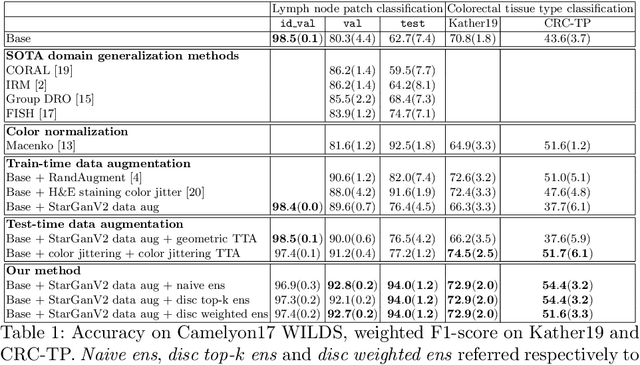
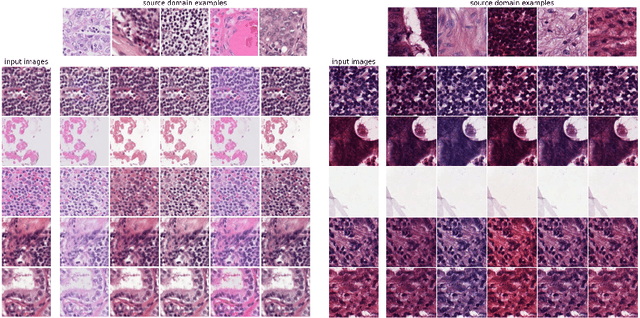
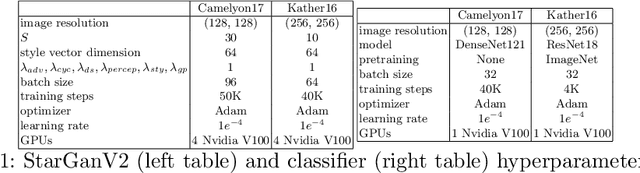
Histopathology whole slide images (WSIs) can reveal significant inter-hospital variability such as illumination, color or optical artifacts. These variations, caused by the use of different scanning protocols across medical centers (staining, scanner), can strongly harm algorithms generalization on unseen protocols. This motivates development of new methods to limit such drop of performances. In this paper, to enhance robustness on unseen target protocols, we propose a new test-time data augmentation based on multi domain image-to-image translation. It allows to project images from unseen protocol into each source domain before classifying them and ensembling the predictions. This test-time augmentation method results in a significant boost of performances for domain generalization. To demonstrate its effectiveness, our method has been evaluated on 2 different histopathology tasks where it outperforms conventional domain generalization, standard H&E specific color augmentation/normalization and standard test-time augmentation techniques. Our code is publicly available at https://gitlab.com/vitadx/articles/test-time-i2i-translation-ensembling.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021Alessa Hering, Lasse Hansen, Tony C. W. Mok, Albert C. S. Chung, Hanna Siebert, Stephanie Häger, Annkristin Lange, Sven Kuckertz, Stefan Heldmann, Wei Shao, Sulaiman Vesal, Mirabela Rusu, Geoffrey Sonn, Théo Estienne, Maria Vakalopoulou, Luyi Han, Yunzhi Huang, Mikael Brudfors, Yaël Balbastre, SamuelJ outard, Marc Modat, Gal Lifshitz, Dan Raviv, Jinxin Lv, Qiang Li, Vincent Jaouen, Dimitris Visvikis, Constance Fourcade, Mathieu Rubeaux, Wentao Pan, Zhe Xu, Bailiang Jian, Francesca De Benetti, Marek Wodzinski, Niklas Gunnarsson, Jens Sjölund, Huaqi Qiu, Zeju Li, Christoph Großbröhmer, Andrew Hoopes, Ingerid Reinertsen, Yiming Xiao, Bennett Landman, Yuankai Huo, Keelin Murphy, Nikolas Lessmann, Bram van Ginneken, Adrian V. Dalca, Mattias P. Heinrich
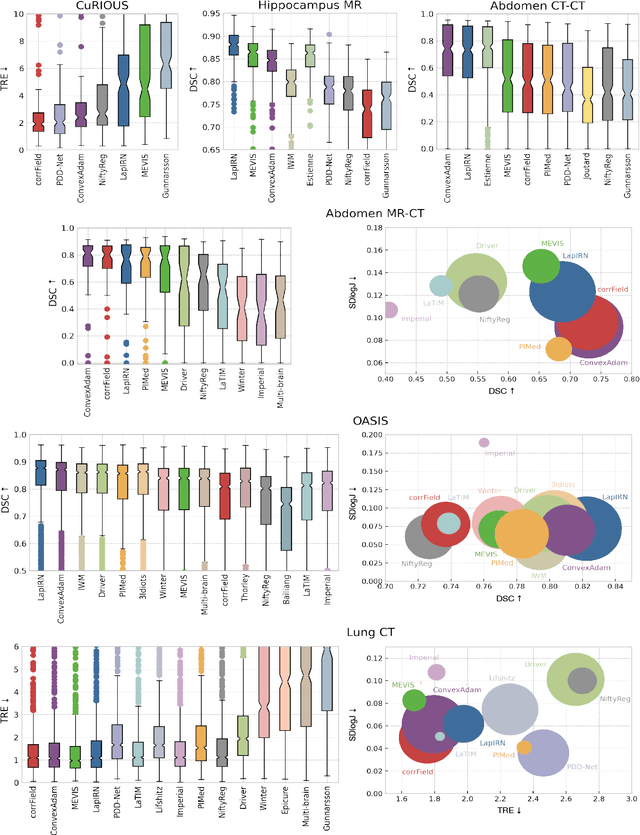
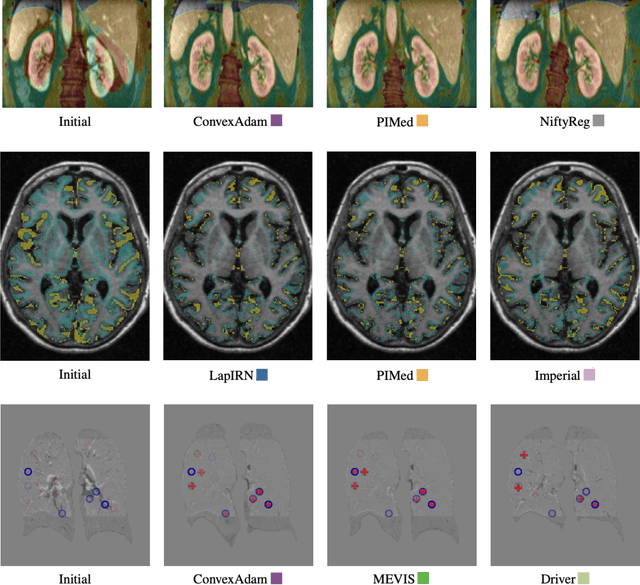
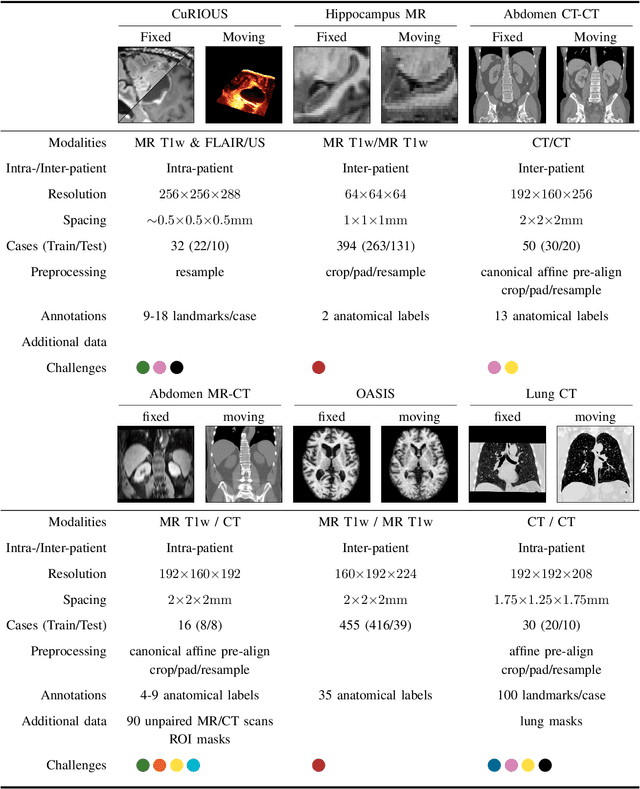
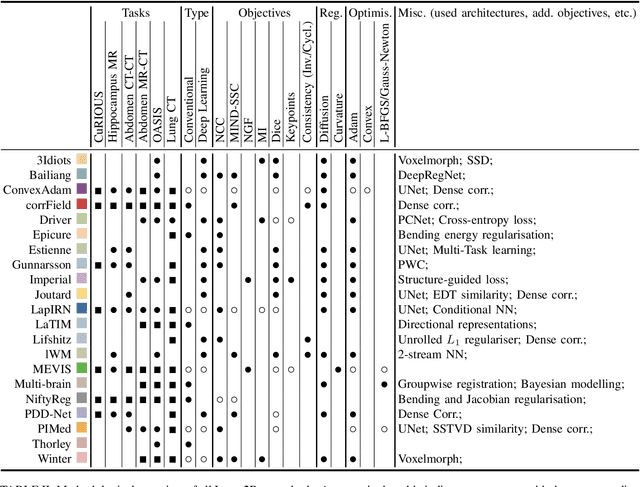
Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge