Vincent Jaouen
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
CT respiratory motion synthesis using joint supervised and adversarial learning
Mar 29, 2024Abstract:Objective: Four-dimensional computed tomography (4DCT) imaging consists in reconstructing a CT acquisition into multiple phases to track internal organ and tumor motion. It is commonly used in radiotherapy treatment planning to establish planning target volumes. However, 4DCT increases protocol complexity, may not align with patient breathing during treatment, and lead to higher radiation delivery. Approach: In this study, we propose a deep synthesis method to generate pseudo respiratory CT phases from static images for motion-aware treatment planning. The model produces patient-specific deformation vector fields (DVFs) by conditioning synthesis on external patient surface-based estimation, mimicking respiratory monitoring devices. A key methodological contribution is to encourage DVF realism through supervised DVF training while using an adversarial term jointly not only on the warped image but also on the magnitude of the DVF itself. This way, we avoid excessive smoothness typically obtained through deep unsupervised learning, and encourage correlations with the respiratory amplitude. Main results: Performance is evaluated using real 4DCT acquisitions with smaller tumor volumes than previously reported. Results demonstrate for the first time that the generated pseudo-respiratory CT phases can capture organ and tumor motion with similar accuracy to repeated 4DCT scans of the same patient. Mean inter-scans tumor center-of-mass distances and Dice similarity coefficients were $1.97$mm and $0.63$, respectively, for real 4DCT phases and $2.35$mm and $0.71$ for synthetic phases, and compares favorably to a state-of-the-art technique (RMSim).
Cross-modal tumor segmentation using generative blending augmentation and self training
Apr 04, 2023Abstract:Deep learning for medical imaging is limited by data scarcity and domain shift, which lead to biased training sets that do not accurately represent deployment conditions. A related practical problem is cross-modal segmentation where the objective is to segment unlabelled domains using previously labelled images from other modalites, which is the context of the MICCAI CrossMoDA 2022 challenge on vestibular schwannoma (VS) segmentation. In this context, we propose a VS segmentation method that leverages conventional image-to-image translation and segmentation using iterative self training combined to a dedicated data augmentation technique called Generative Blending Augmentation (GBA). GBA is based on a one-shot 2D SinGAN generative model that allows to realistically diversify target tumor appearances in a downstream segmentation model, improving its generalization power at test time. Our solution ranked first on the VS segmentation task during the validation and test phase of the CrossModa 2022 challenge.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021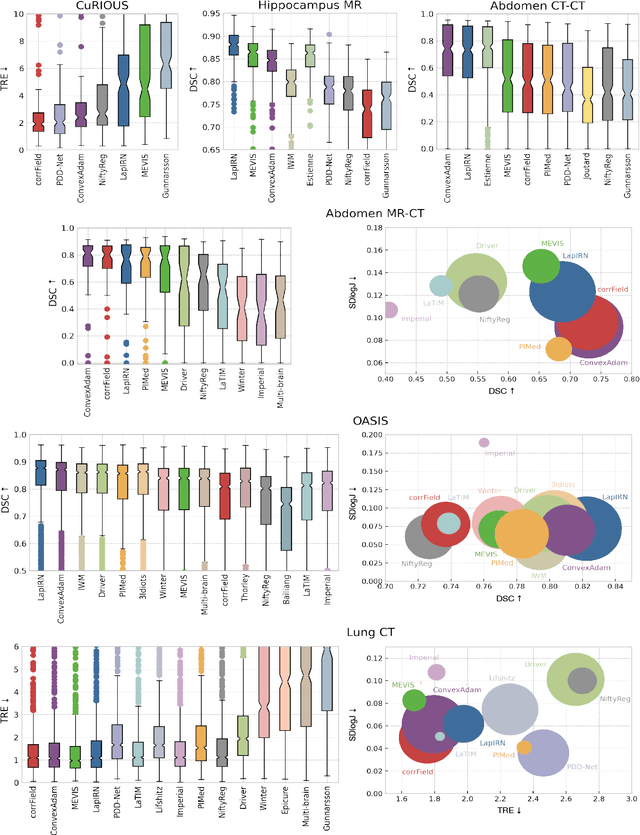
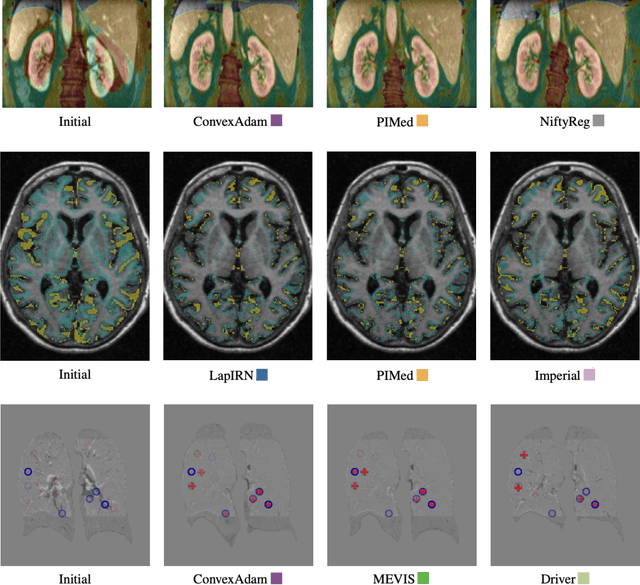
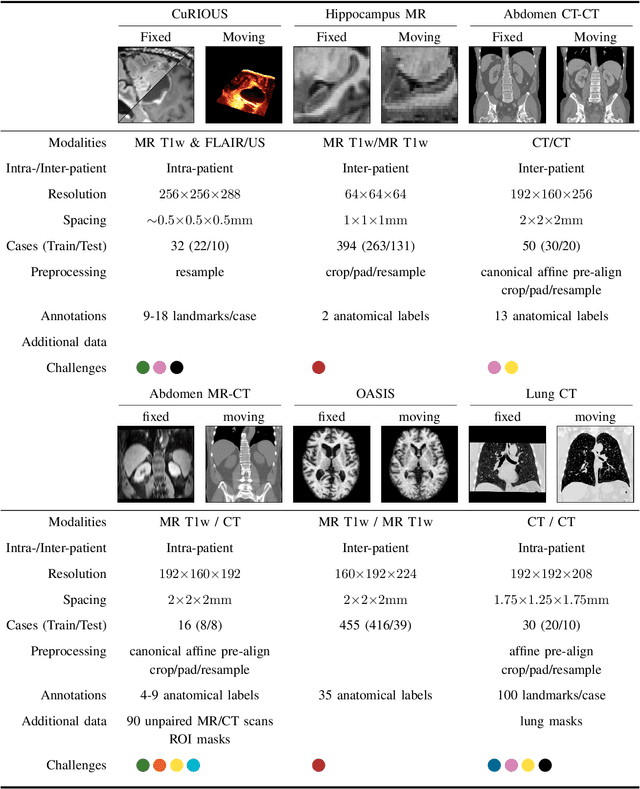
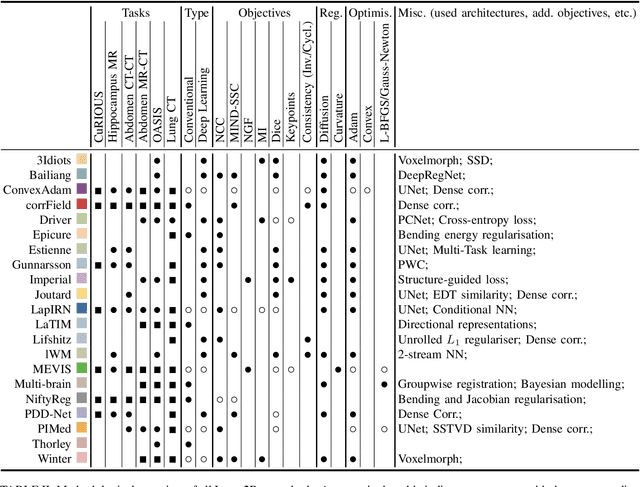
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Regularized directional representations for medical image registration
Nov 30, 2021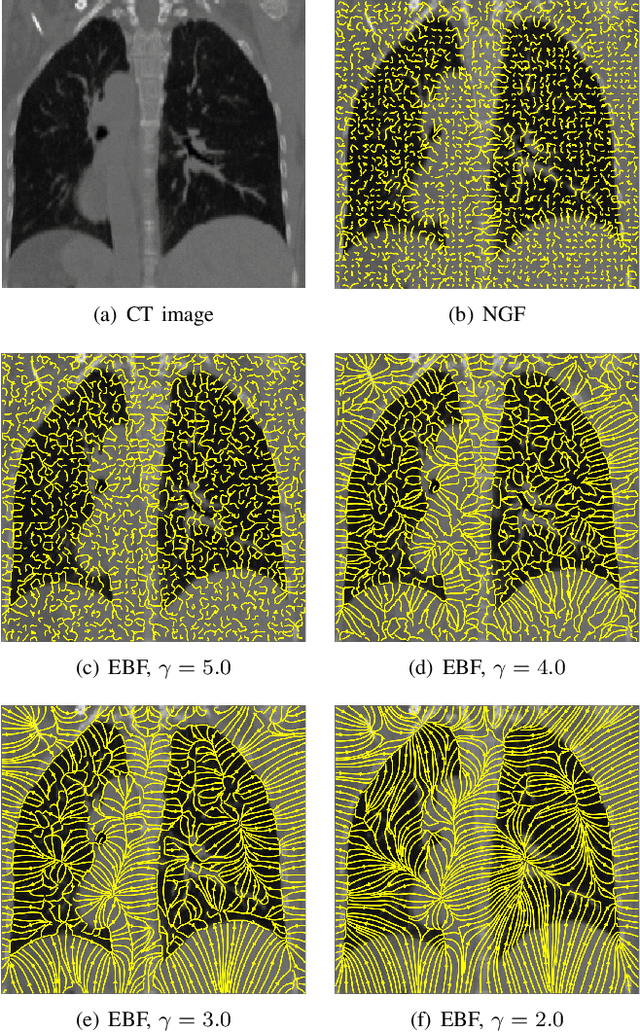
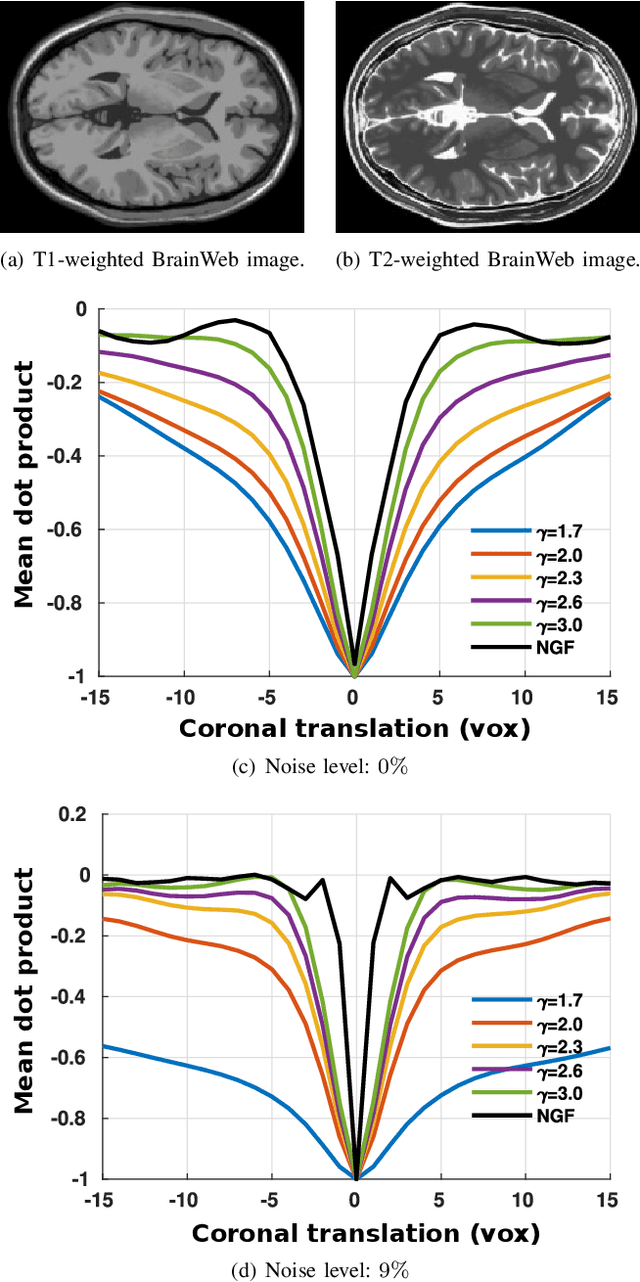
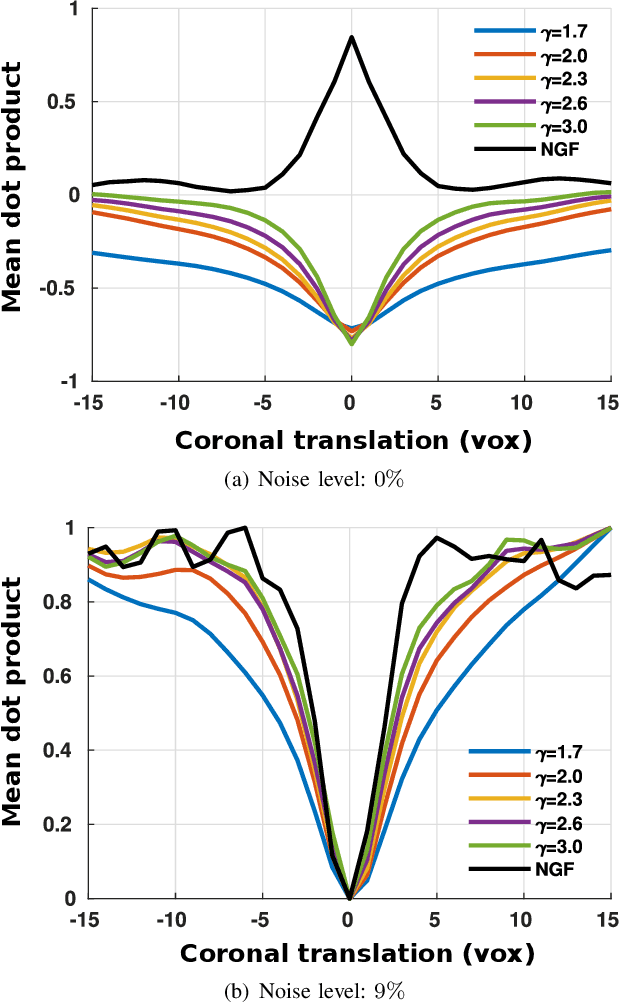
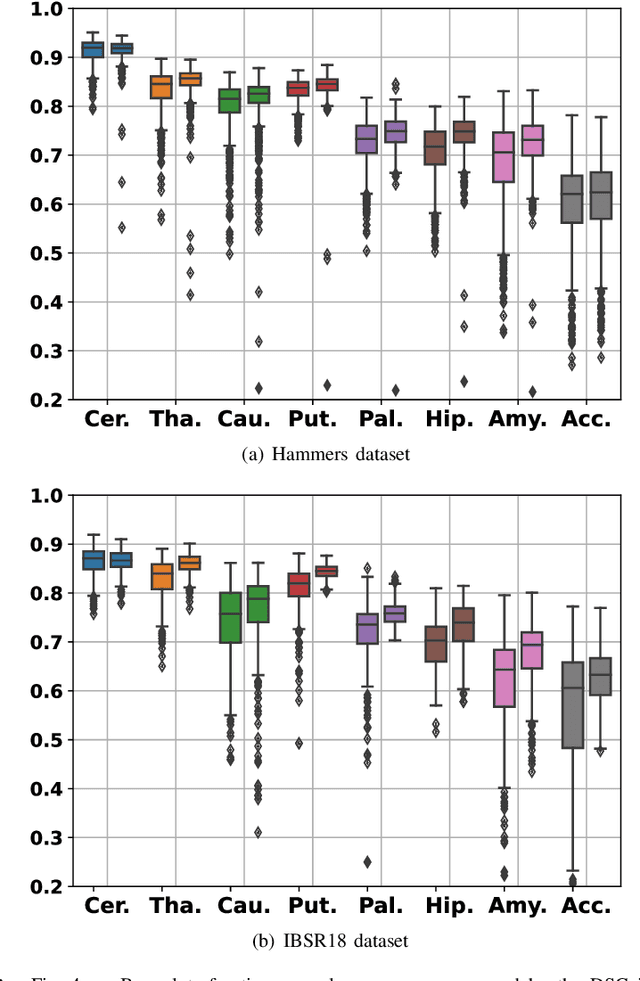
Abstract:In image registration, many efforts have been devoted to the development of alternatives to the popular normalized mutual information criterion. Concurrently to these efforts, an increasing number of works have demonstrated that substantial gains in registration accuracy can also be achieved by aligning structural representations of images rather than images themselves. Following this research path, we propose a new method for mono- and multimodal image registration based on the alignment of regularized vector fields derived from structural information such as gradient vector flow fields, a technique we call \textit{vector field similarity}. Our approach can be combined in a straightforward fashion with any existing registration framework by substituting vector field similarity to intensity-based registration. In our experiments, we show that the proposed approach compares favourably with conventional image alignment on several public image datasets using a diversity of imaging modalities and anatomical locations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge