Jinxin Lv
Robust One-shot Segmentation of Brain Tissues via Image-aligned Style Transformation
Nov 30, 2022



Abstract:One-shot segmentation of brain tissues is typically a dual-model iterative learning: a registration model (reg-model) warps a carefully-labeled atlas onto unlabeled images to initialize their pseudo masks for training a segmentation model (seg-model); the seg-model revises the pseudo masks to enhance the reg-model for a better warping in the next iteration. However, there is a key weakness in such dual-model iteration that the spatial misalignment inevitably caused by the reg-model could misguide the seg-model, which makes it converge on an inferior segmentation performance eventually. In this paper, we propose a novel image-aligned style transformation to reinforce the dual-model iterative learning for robust one-shot segmentation of brain tissues. Specifically, we first utilize the reg-model to warp the atlas onto an unlabeled image, and then employ the Fourier-based amplitude exchange with perturbation to transplant the style of the unlabeled image into the aligned atlas. This allows the subsequent seg-model to learn on the aligned and style-transferred copies of the atlas instead of unlabeled images, which naturally guarantees the correct spatial correspondence of an image-mask training pair, without sacrificing the diversity of intensity patterns carried by the unlabeled images. Furthermore, we introduce a feature-aware content consistency in addition to the image-level similarity to constrain the reg-model for a promising initialization, which avoids the collapse of image-aligned style transformation in the first iteration. Experimental results on two public datasets demonstrate 1) a competitive segmentation performance of our method compared to the fully-supervised method, and 2) a superior performance over other state-of-the-art with an increase of average Dice by up to 4.67%. The source code is available at: https://github.com/JinxLv/One-shot-segmentation-via-IST.
Accurate Scoliosis Vertebral Landmark Localization on X-ray Images via Shape-constrained Multi-stage Cascaded CNNs
Jun 05, 2022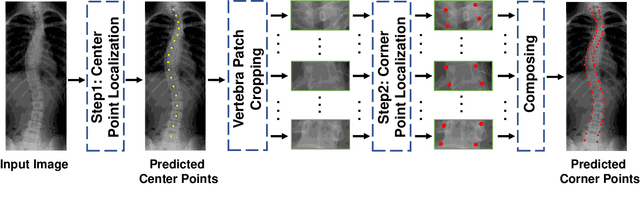
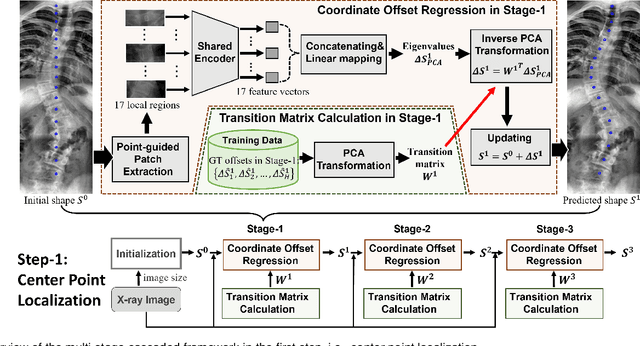
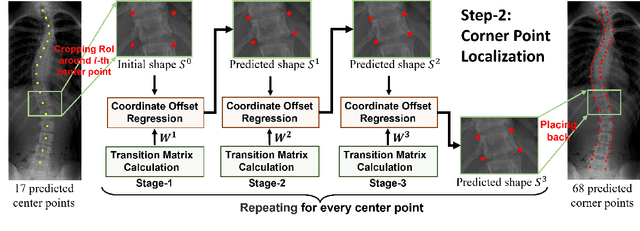
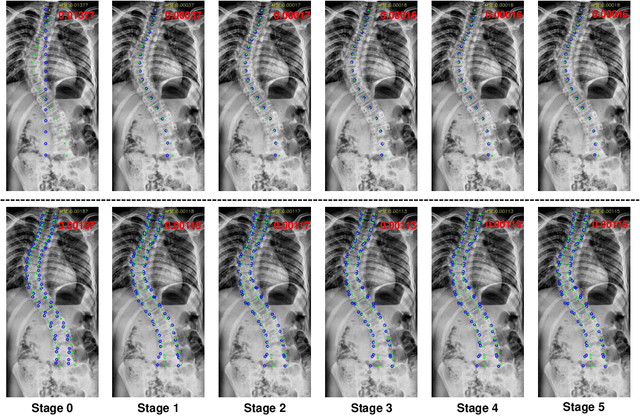
Abstract:Vertebral landmark localization is a crucial step for variant spine-related clinical applications, which requires detecting the corner points of 17 vertebrae. However, the neighbor landmarks often disturb each other for the homogeneous appearance of vertebrae, which makes vertebral landmark localization extremely difficult. In this paper, we propose multi-stage cascaded convolutional neural networks (CNNs) to split the single task into two sequential steps, i.e., center point localization to roughly locate 17 center points of vertebrae, and corner point localization to find 4 corner points for each vertebra without distracted by others. Landmarks in each step are located gradually from a set of initialized points by regressing offsets via cascaded CNNs. Principal Component Analysis (PCA) is employed to preserve a shape constraint in offset regression to resist the mutual attraction of vertebrae. We evaluate our method on the AASCE dataset that consists of 609 tight spinal anterior-posterior X-ray images and each image contains 17 vertebrae composed of the thoracic and lumbar spine for spinal shape characterization. Experimental results demonstrate our superior performance of vertebral landmark localization over other state-of-the-arts with the relative error decreasing from 3.2e-3 to 7.2e-4.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021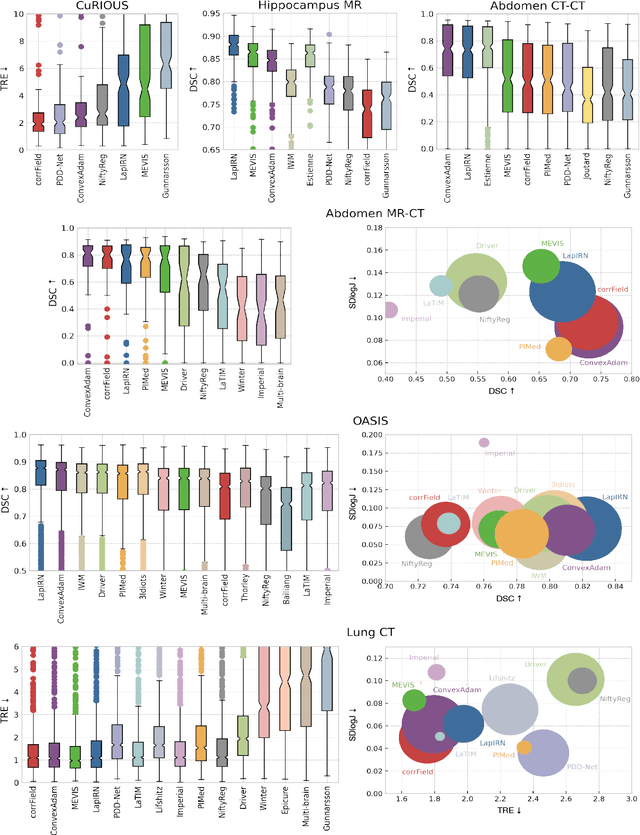
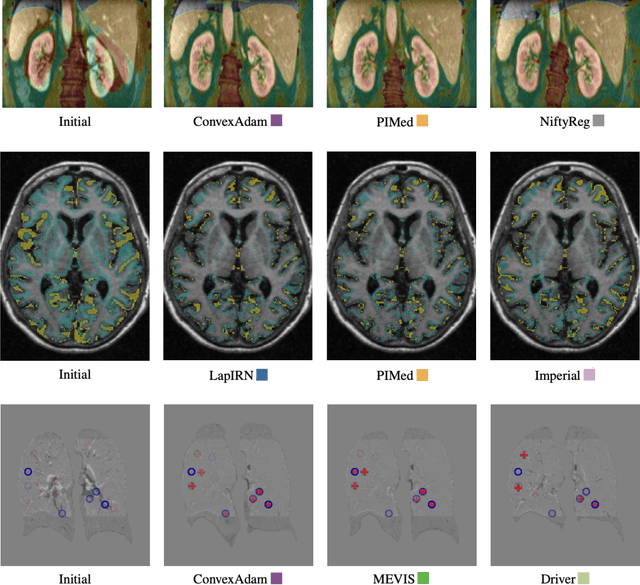
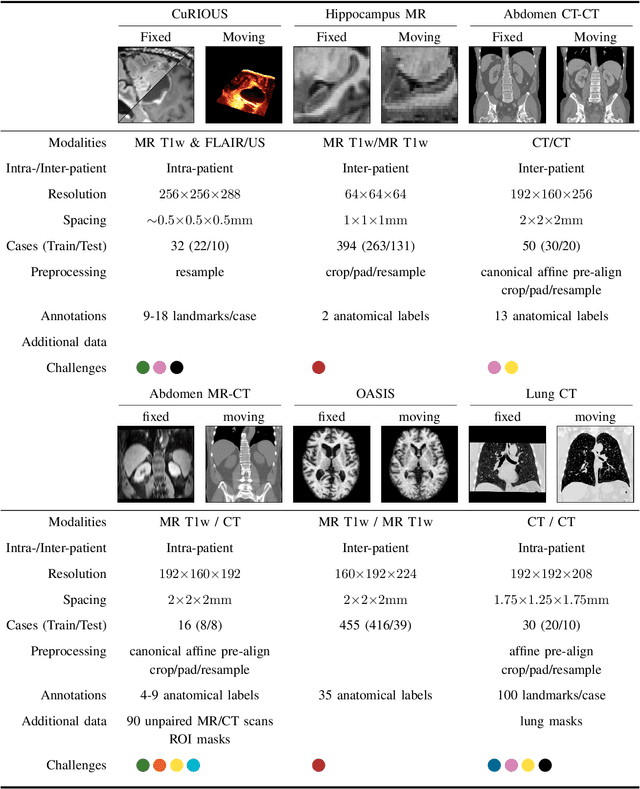
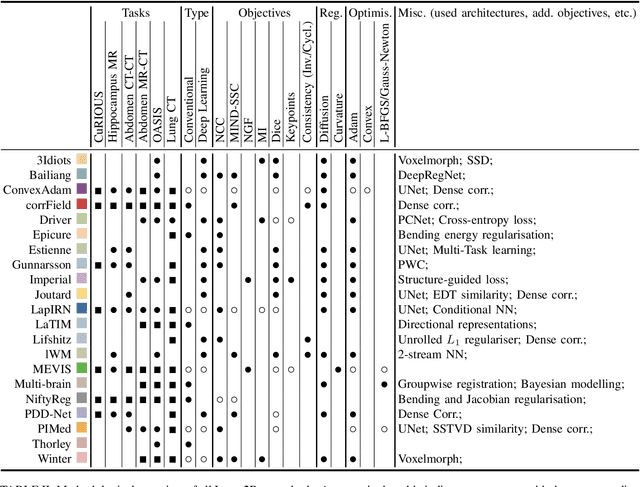
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Joint Progressive and Coarse-to-fine Registration of Brain MRI via Deformation Field Integration and Non-Rigid Feature Fusion
Sep 25, 2021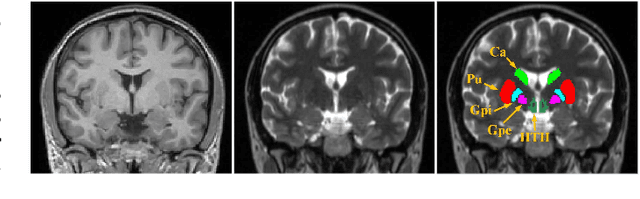
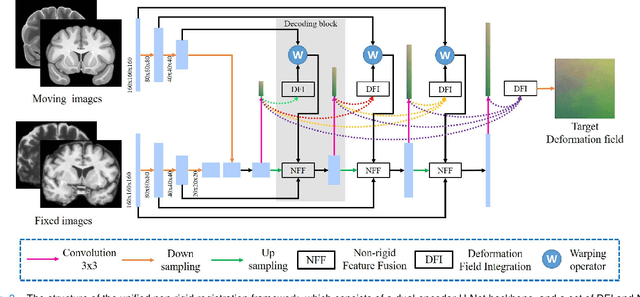
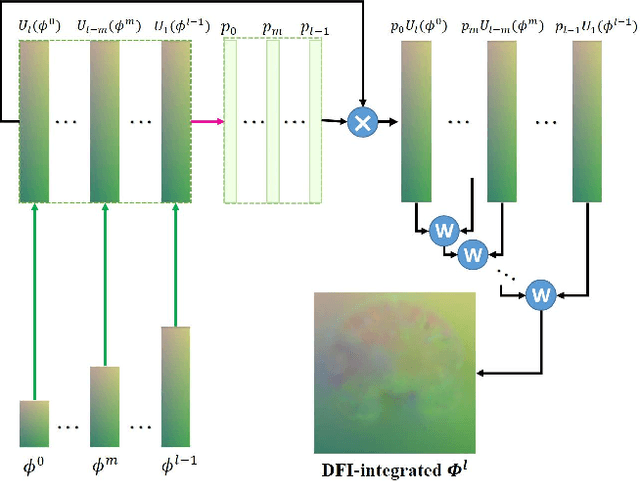
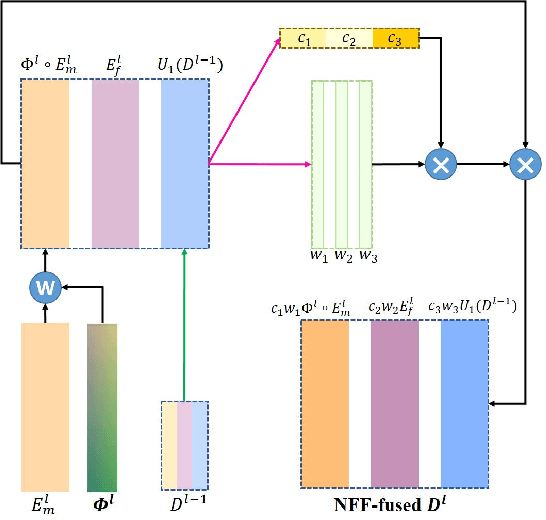
Abstract:Registration of brain MRI images requires to solve a deformation field, which is extremely difficult in aligning intricate brain tissues, e.g., subcortical nuclei, etc. Existing efforts resort to decomposing the target deformation field into intermediate sub-fields with either tiny motions, i.e., progressive registration stage by stage, or lower resolutions, i.e., coarse-to-fine estimation of the full-size deformation field. In this paper, we argue that those efforts are not mutually exclusive, and propose a unified framework for robust brain MRI registration in both progressive and coarse-to-fine manners simultaneously. Specifically, building on a dual-encoder U-Net, the fixed-moving MRI pair is encoded and decoded into multi-scale deformation sub-fields from coarse to fine. Each decoding block contains two proposed novel modules: i) in Deformation Field Integration (DFI), a single integrated sub-field is calculated, warping by which is equivalent to warping progressively by sub-fields from all previous decoding blocks, and ii) in Non-rigid Feature Fusion (NFF), features of the fixed-moving pair are aligned by DFI-integrated sub-field, and then fused to predict a finer sub-field. Leveraging both DFI and NFF, the target deformation field is factorized into multi-scale sub-fields, where the coarser fields alleviate the estimate of a finer one and the finer field learns to make up those misalignments insolvable by previous coarser ones. The extensive and comprehensive experimental results on both private and public datasets demonstrate a superior registration performance of brain MRI images over progressive registration only and coarse-to-fine estimation only, with an increase by at most 10% in the average Dice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge