Andrew Hoopes
Unified Brain Surface and Volume Registration
Dec 22, 2025Abstract:Accurate registration of brain MRI scans is fundamental for cross-subject analysis in neuroscientific studies. This involves aligning both the cortical surface of the brain and the interior volume. Traditional methods treat volumetric and surface-based registration separately, which often leads to inconsistencies that limit downstream analyses. We propose a deep learning framework, NeurAlign, that registers $3$D brain MRI images by jointly aligning both cortical and subcortical regions through a unified volume-and-surface-based representation. Our approach leverages an intermediate spherical coordinate space to bridge anatomical surface topology with volumetric anatomy, enabling consistent and anatomically accurate alignment. By integrating spherical registration into the learning, our method ensures geometric coherence between volume and surface domains. In a series of experiments on both in-domain and out-of-domain datasets, our method consistently outperforms both classical and machine learning-based registration methods -- improving the Dice score by up to 7 points while maintaining regular deformation fields. Additionally, it is orders of magnitude faster than the standard method for this task, and is simpler to use because it requires no additional inputs beyond an MRI scan. With its superior accuracy, fast inference, and ease of use, NeurAlign sets a new standard for joint cortical and subcortical registration.
AtlasMorph: Learning conditional deformable templates for brain MRI
Nov 17, 2025



Abstract:Deformable templates, or atlases, are images that represent a prototypical anatomy for a population, and are often enhanced with probabilistic anatomical label maps. They are commonly used in medical image analysis for population studies and computational anatomy tasks such as registration and segmentation. Because developing a template is a computationally expensive process, relatively few templates are available. As a result, analysis is often conducted with sub-optimal templates that are not truly representative of the study population, especially when there are large variations within this population. We propose a machine learning framework that uses convolutional registration neural networks to efficiently learn a function that outputs templates conditioned on subject-specific attributes, such as age and sex. We also leverage segmentations, when available, to produce anatomical segmentation maps for the resulting templates. The learned network can also be used to register subject images to the templates. We demonstrate our method on a compilation of 3D brain MRI datasets, and show that it can learn high-quality templates that are representative of populations. We find that annotated conditional templates enable better registration than their unlabeled unconditional counterparts, and outperform other templates construction methods.
End-to-end Cortical Surface Reconstruction from Clinical Magnetic Resonance Images
May 20, 2025Abstract:Surface-based cortical analysis is valuable for a variety of neuroimaging tasks, such as spatial normalization, parcellation, and gray matter (GM) thickness estimation. However, most tools for estimating cortical surfaces work exclusively on scans with at least 1 mm isotropic resolution and are tuned to a specific magnetic resonance (MR) contrast, often T1-weighted (T1w). This precludes application using most clinical MR scans, which are very heterogeneous in terms of contrast and resolution. Here, we use synthetic domain-randomized data to train the first neural network for explicit estimation of cortical surfaces from scans of any contrast and resolution, without retraining. Our method deforms a template mesh to the white matter (WM) surface, which guarantees topological correctness. This mesh is further deformed to estimate the GM surface. We compare our method to recon-all-clinical (RAC), an implicit surface reconstruction method which is currently the only other tool capable of processing heterogeneous clinical MR scans, on ADNI and a large clinical dataset (n=1,332). We show a approximately 50 % reduction in cortical thickness error (from 0.50 to 0.24 mm) with respect to RAC and better recovery of the aging-related cortical thinning patterns detected by FreeSurfer on high-resolution T1w scans. Our method enables fast and accurate surface reconstruction of clinical scans, allowing studies (1) with sample sizes far beyond what is feasible in a research setting, and (2) of clinical populations that are difficult to enroll in research studies. The code is publicly available at https://github.com/simnibs/brainnet.
MultiMorph: On-demand Atlas Construction
Mar 31, 2025Abstract:We present MultiMorph, a fast and efficient method for constructing anatomical atlases on the fly. Atlases capture the canonical structure of a collection of images and are essential for quantifying anatomical variability across populations. However, current atlas construction methods often require days to weeks of computation, thereby discouraging rapid experimentation. As a result, many scientific studies rely on suboptimal, precomputed atlases from mismatched populations, negatively impacting downstream analyses. MultiMorph addresses these challenges with a feedforward model that rapidly produces high-quality, population-specific atlases in a single forward pass for any 3D brain dataset, without any fine-tuning or optimization. MultiMorph is based on a linear group-interaction layer that aggregates and shares features within the group of input images. Further, by leveraging auxiliary synthetic data, MultiMorph generalizes to new imaging modalities and population groups at test-time. Experimentally, MultiMorph outperforms state-of-the-art optimization-based and learning-based atlas construction methods in both small and large population settings, with a 100-fold reduction in time. This makes MultiMorph an accessible framework for biomedical researchers without machine learning expertise, enabling rapid, high-quality atlas generation for diverse studies.
VoxelPrompt: A Vision-Language Agent for Grounded Medical Image Analysis
Oct 10, 2024



Abstract:We present VoxelPrompt, an agent-driven vision-language framework that tackles diverse radiological tasks through joint modeling of natural language, image volumes, and analytical metrics. VoxelPrompt is multi-modal and versatile, leveraging the flexibility of language interaction while providing quantitatively grounded image analysis. Given a variable number of 3D medical volumes, such as MRI and CT scans, VoxelPrompt employs a language agent that iteratively predicts executable instructions to solve a task specified by an input prompt. These instructions communicate with a vision network to encode image features and generate volumetric outputs (e.g., segmentations). VoxelPrompt interprets the results of intermediate instructions and plans further actions to compute discrete measures (e.g., tumor growth across a series of scans) and present relevant outputs to the user. We evaluate this framework in a sandbox of diverse neuroimaging tasks, and we show that the single VoxelPrompt model can delineate hundreds of anatomical and pathological features, measure many complex morphological properties, and perform open-language analysis of lesion characteristics. VoxelPrompt carries out these objectives with accuracy similar to that of fine-tuned, single-task models for segmentation and visual question-answering, while facilitating a much larger range of tasks. Therefore, by supporting accurate image processing with language interaction, VoxelPrompt provides comprehensive utility for numerous imaging tasks that traditionally require specialized models to address.
Anatomy-aware and acquisition-agnostic joint registration with SynthMorph
Jan 26, 2023Abstract:Affine image registration is a cornerstone of medical-image processing and analysis. While classical algorithms can achieve excellent accuracy, they solve a time-consuming optimization for every new image pair. Deep-learning (DL) methods learn a function that maps an image pair to an output transform. Evaluating the functions is fast, but capturing large transforms can be challenging, and networks tend to struggle if a test-image characteristic shifts from the training domain, such as the contrast or resolution. A majority of affine methods are also agnostic to the anatomy the user wishes to align; the registration will be inaccurate if algorithms consider all structures in the image. We address these shortcomings with a fast, robust, and easy-to-use DL tool for affine and deformable registration of any brain image without preprocessing, right off the MRI scanner. First, we rigorously analyze how competing architectures learn affine transforms across a diverse set of neuroimaging data, aiming to truly capture the behavior of methods in the real world. Second, we leverage a recent strategy to train networks with wildly varying images synthesized from label maps, yielding robust performance across acquisition specifics. Third, we optimize the spatial overlap of select anatomical labels, which enables networks to distinguish between anatomy of interest and irrelevant structures, removing the need for preprocessing that excludes content that would otherwise reduce the accuracy of anatomy-specific registration. We combine the affine model with prior work on deformable registration and test brain-specific registration across a landscape of MRI protocols unseen at training, demonstrating consistent and improved accuracy compared to existing tools. We distribute our code and tool at https://w3id.org/synthmorph, providing a single complete end-to-end solution for registration of brain MRI.
An Open-Source Tool for Longitudinal Whole-Brain and White Matter Lesion Segmentation
Jul 10, 2022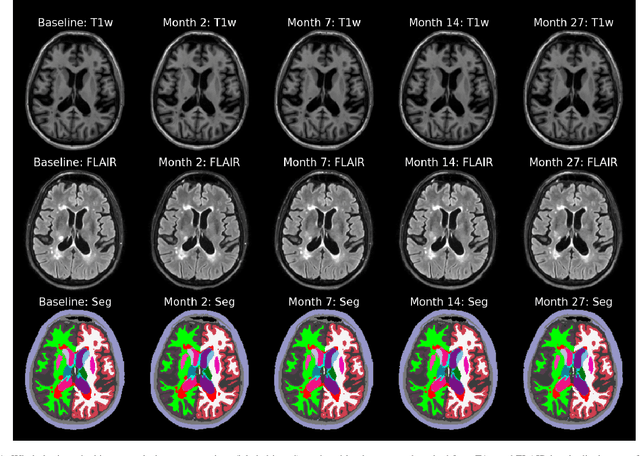
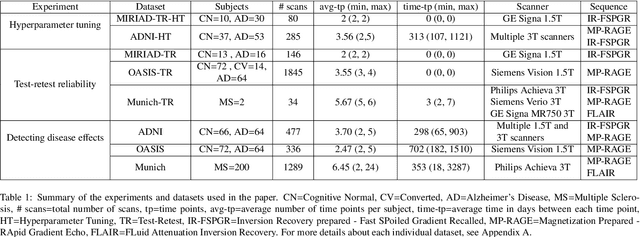
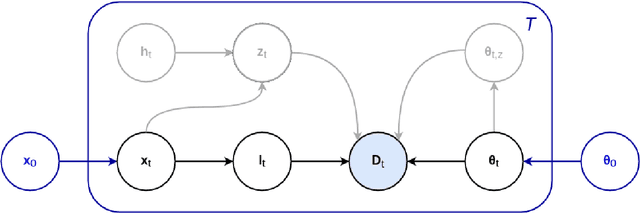

Abstract:In this paper we describe and validate a longitudinal method for whole-brain segmentation of longitudinal MRI scans. It builds upon an existing whole-brain segmentation method that can handle multi-contrast data and robustly analyze images with white matter lesions. This method is here extended with subject-specific latent variables that encourage temporal consistency between its segmentation results, enabling it to better track subtle morphological changes in dozens of neuroanatomical structures and white matter lesions. We validate the proposed method on multiple datasets of control subjects and patients suffering from Alzheimer's disease and multiple sclerosis, and compare its results against those obtained with its original cross-sectional formulation and two benchmark longitudinal methods. The results indicate that the method attains a higher test-retest reliability, while being more sensitive to longitudinal disease effect differences between patient groups. An implementation is publicly available as part of the open-source neuroimaging package FreeSurfer.
Learning the Effect of Registration Hyperparameters with HyperMorph
Mar 30, 2022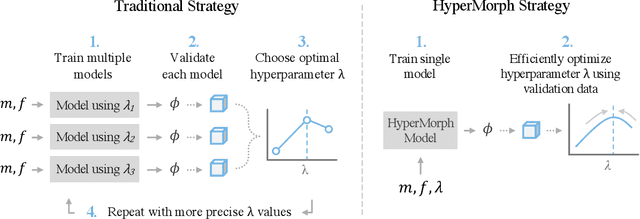
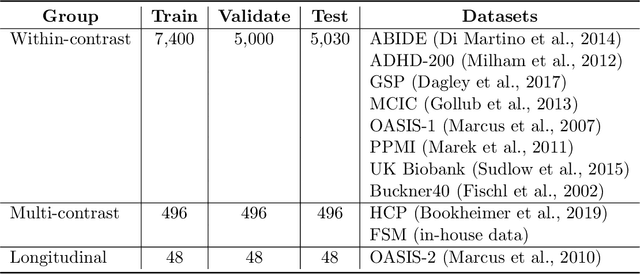

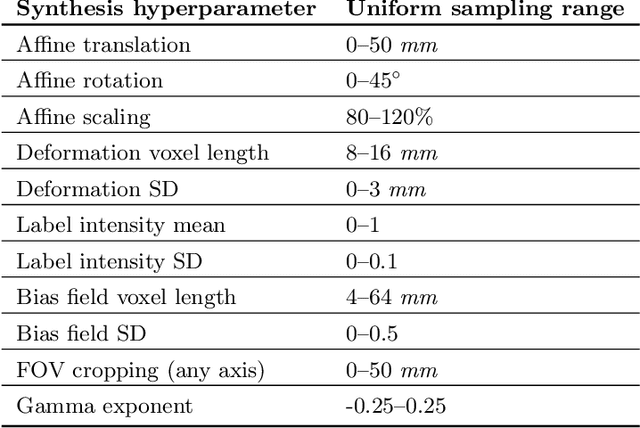
Abstract:We introduce HyperMorph, a framework that facilitates efficient hyperparameter tuning in learning-based deformable image registration. Classical registration algorithms perform an iterative pair-wise optimization to compute a deformation field that aligns two images. Recent learning-based approaches leverage large image datasets to learn a function that rapidly estimates a deformation for a given image pair. In both strategies, the accuracy of the resulting spatial correspondences is strongly influenced by the choice of certain hyperparameter values. However, an effective hyperparameter search consumes substantial time and human effort as it often involves training multiple models for different fixed hyperparameter values and may lead to suboptimal registration. We propose an amortized hyperparameter learning strategy to alleviate this burden by learning the impact of hyperparameters on deformation fields. We design a meta network, or hypernetwork, that predicts the parameters of a registration network for input hyperparameters, thereby comprising a single model that generates the optimal deformation field corresponding to given hyperparameter values. This strategy enables fast, high-resolution hyperparameter search at test-time, reducing the inefficiency of traditional approaches while increasing flexibility. We also demonstrate additional benefits of HyperMorph, including enhanced robustness to model initialization and the ability to rapidly identify optimal hyperparameter values specific to a dataset, image contrast, task, or even anatomical region, all without the need to retrain models. We make our code publicly available at http://hypermorph.voxelmorph.net.
SynthStrip: Skull-Stripping for Any Brain Image
Mar 18, 2022
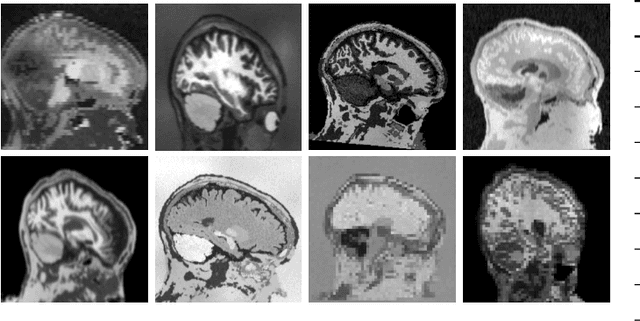
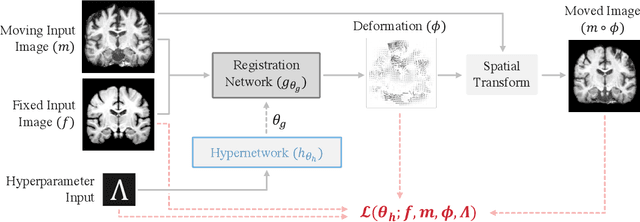
Abstract:The removal of non-brain signal from magnetic resonance imaging (MRI) data, known as skull-stripping, is an integral component of many neuroimage analysis streams. Despite their abundance, popular classical skull-stripping methods are usually tailored to images with specific acquisition properties, namely near-isotropic resolution and T1-weighted (T1w) MRI contrast, which are prevalent in research settings. As a result, existing tools tend to adapt poorly to other image types, such as stacks of thick slices acquired with fast spin-echo (FSE) MRI that are common in the clinic. While learning-based approaches for brain extraction have gained traction in recent years, these methods face a similar burden, as they are only effective for image types seen during the training procedure. To achieve robust skull-stripping across a landscape of protocols, we introduce SynthStrip, a rapid, learning-based brain-extraction tool. By leveraging anatomical segmentations to generate an entirely synthetic training dataset with anatomies, intensity distributions, and artifacts that far exceed the realistic range of medical images, SynthStrip learns to successfully generalize to a variety of real acquired brain images, removing the need for training data with target contrasts. We demonstrate the efficacy of SynthStrip for a diverse set of image acquisitions and resolutions across subject populations, ranging from newborn to adult. We show substantial improvements in accuracy over popular skull-stripping baselines - all with a single trained model. Our method and labeled evaluation data are available at https://w3id.org/synthstrip.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021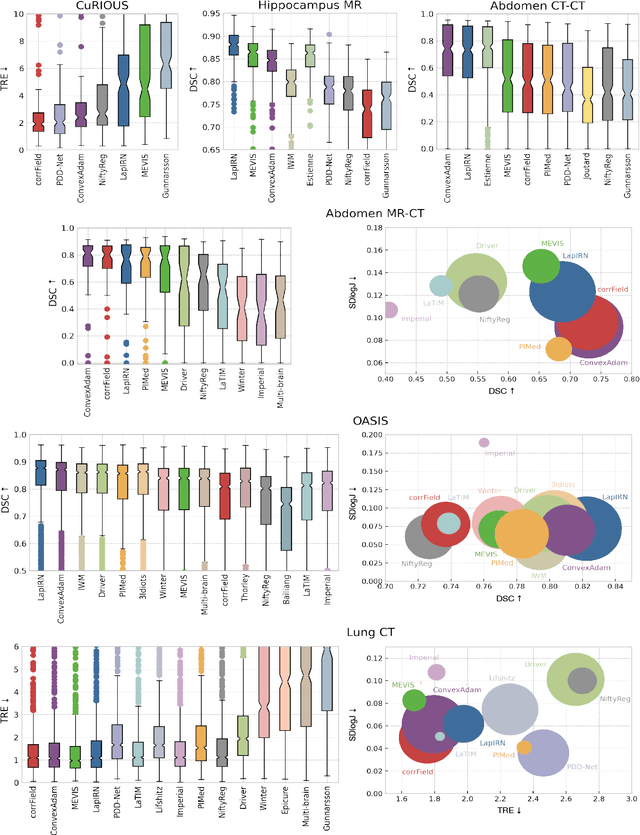
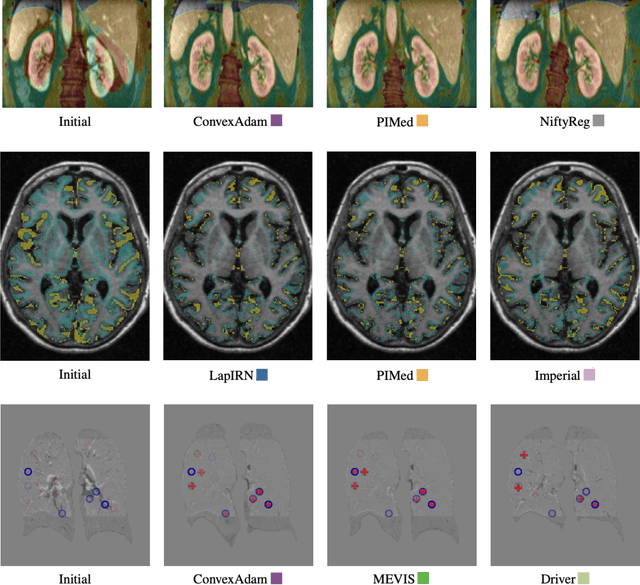
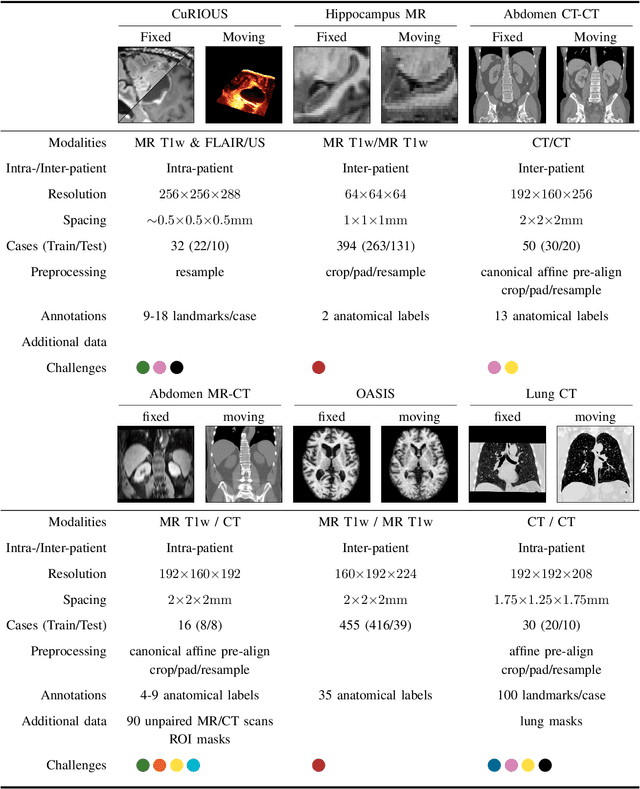
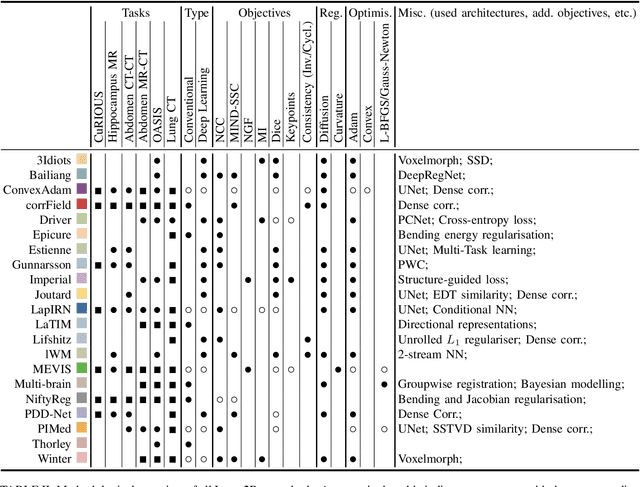
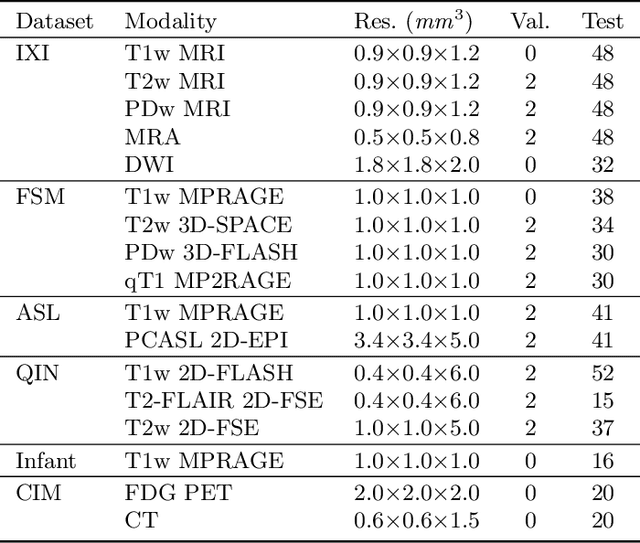
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge