Mattias P. Heinrich
Depth to Anatomy: Learning Internal Organ Locations from Surface Depth Images
Jan 26, 2026Abstract:Automated patient positioning plays an important role in optimizing scanning procedure and improving patient throughput. Leveraging depth information captured by RGB-D cameras presents a promising approach for estimating internal organ positions, thereby enabling more accurate and efficient positioning. In this work, we propose a learning-based framework that directly predicts the 3D locations and shapes of multiple internal organs from single 2D depth images of the body surface. Utilizing a large-scale dataset of full-body MRI scans, we synthesize depth images paired with corresponding anatomical segmentations to train a unified convolutional neural network architecture. Our method accurately localizes a diverse set of anatomical structures, including bones and soft tissues, without requiring explicit surface reconstruction. Experimental results demonstrate the potential of integrating depth sensors into radiology workflows to streamline scanning procedures and enhance patient experience through automated patient positioning.
Fracture Morphology Classification: Local Multiclass Modeling for Multilabel Complexity
Dec 16, 2025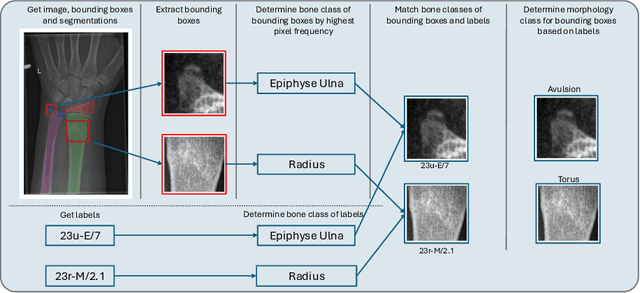
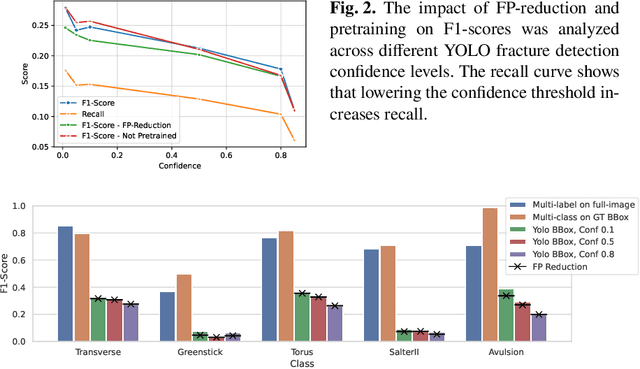
Abstract:Between $15\,\%$ and $45\,\%$ of children experience a fracture during their growth years, making accurate diagnosis essential. Fracture morphology, alongside location and fragment angle, is a key diagnostic feature. In this work, we propose a method to extract fracture morphology by assigning automatically global AO codes to corresponding fracture bounding boxes. This approach enables the use of public datasets and reformulates the global multilabel task into a local multiclass one, improving the average F1 score by $7.89\,\%$. However, performance declines when using imperfect fracture detectors, highlighting challenges for real-world deployment. Our code is available on GitHub.
PrIINeR: Towards Prior-Informed Implicit Neural Representations for Accelerated MRI
Aug 11, 2025Abstract:Accelerating Magnetic Resonance Imaging (MRI) reduces scan time but often degrades image quality. While Implicit Neural Representations (INRs) show promise for MRI reconstruction, they struggle at high acceleration factors due to weak prior constraints, leading to structural loss and aliasing artefacts. To address this, we propose PrIINeR, an INR-based MRI reconstruction method that integrates prior knowledge from pre-trained deep learning models into the INR framework. By combining population-level knowledge with instance-based optimization and enforcing dual data consistency, PrIINeR aligns both with the acquired k-space data and the prior-informed reconstruction. Evaluated on the NYU fastMRI dataset, our method not only outperforms state-of-the-art INR-based approaches but also improves upon several learning-based state-of-the-art methods, significantly improving structural preservation and fidelity while effectively removing aliasing artefacts.PrIINeR bridges deep learning and INR-based techniques, offering a more reliable solution for high-quality, accelerated MRI reconstruction. The code is publicly available on https://github.com/multimodallearning/PrIINeR.
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025



Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
OncoReg: Medical Image Registration for Oncological Challenges
Apr 01, 2025Abstract:In modern cancer research, the vast volume of medical data generated is often underutilised due to challenges related to patient privacy. The OncoReg Challenge addresses this issue by enabling researchers to develop and validate image registration methods through a two-phase framework that ensures patient privacy while fostering the development of more generalisable AI models. Phase one involves working with a publicly available dataset, while phase two focuses on training models on a private dataset within secure hospital networks. OncoReg builds upon the foundation established by the Learn2Reg Challenge by incorporating the registration of interventional cone-beam computed tomography (CBCT) with standard planning fan-beam CT (FBCT) images in radiotherapy. Accurate image registration is crucial in oncology, particularly for dynamic treatment adjustments in image-guided radiotherapy, where precise alignment is necessary to minimise radiation exposure to healthy tissues while effectively targeting tumours. This work details the methodology and data behind the OncoReg Challenge and provides a comprehensive analysis of the competition entries and results. Findings reveal that feature extraction plays a pivotal role in this registration task. A new method emerging from this challenge demonstrated its versatility, while established approaches continue to perform comparably to newer techniques. Both deep learning and classical approaches still play significant roles in image registration, with the combination of methods - particularly in feature extraction - proving most effective.
Internal Organ Localization Using Depth Images
Mar 30, 2025Abstract:Automated patient positioning is a crucial step in streamlining MRI workflows and enhancing patient throughput. RGB-D camera-based systems offer a promising approach to automate this process by leveraging depth information to estimate internal organ positions. This paper investigates the feasibility of a learning-based framework to infer approximate internal organ positions from the body surface. Our approach utilizes a large-scale dataset of MRI scans to train a deep learning model capable of accurately predicting organ positions and shapes from depth images alone. We demonstrate the effectiveness of our method in localization of multiple internal organs, including bones and soft tissues. Our findings suggest that RGB-D camera-based systems integrated into MRI workflows have the potential to streamline scanning procedures and improve patient experience by enabling accurate and automated patient positioning.
PULPo: Probabilistic Unsupervised Laplacian Pyramid Registration
Jul 15, 2024



Abstract:Deformable image registration is fundamental to many medical imaging applications. Registration is an inherently ambiguous task often admitting many viable solutions. While neural network-based registration techniques enable fast and accurate registration, the majority of existing approaches are not able to estimate uncertainty. Here, we present PULPo, a method for probabilistic deformable registration capable of uncertainty quantification. PULPo probabilistically models the distribution of deformation fields on different hierarchical levels combining them using Laplacian pyramids. This allows our method to model global as well as local aspects of the deformation field. We evaluate our method on two widely used neuroimaging datasets and find that it achieves high registration performance as well as substantially better calibrated uncertainty quantification compared to the current state-of-the-art.
IM-MoCo: Self-supervised MRI Motion Correction using Motion-Guided Implicit Neural Representations
Jul 03, 2024Abstract:Motion artifacts in Magnetic Resonance Imaging (MRI) arise due to relatively long acquisition times and can compromise the clinical utility of acquired images. Traditional motion correction methods often fail to address severe motion, leading to distorted and unreliable results. Deep Learning (DL) alleviated such pitfalls through generalization with the cost of vanishing structures and hallucinations, making it challenging to apply in the medical field where hallucinated structures can tremendously impact the diagnostic outcome. In this work, we present an instance-wise motion correction pipeline that leverages motion-guided Implicit Neural Representations (INRs) to mitigate the impact of motion artifacts while retaining anatomical structure. Our method is evaluated using the NYU fastMRI dataset with different degrees of simulated motion severity. For the correction alone, we can improve over state-of-the-art image reconstruction methods by $+5\%$ SSIM, $+5\:db$ PSNR, and $+14\%$ HaarPSI. Clinical relevance is demonstrated by a subsequent experiment, where our method improves classification outcomes by at least $+1.5$ accuracy percentage points compared to motion-corrupted images.
Self-supervised Learning of Dense Hierarchical Representations for Medical Image Segmentation
Jan 12, 2024



Abstract:This paper demonstrates a self-supervised framework for learning voxel-wise coarse-to-fine representations tailored for dense downstream tasks. Our approach stems from the observation that existing methods for hierarchical representation learning tend to prioritize global features over local features due to inherent architectural bias. To address this challenge, we devise a training strategy that balances the contributions of features from multiple scales, ensuring that the learned representations capture both coarse and fine-grained details. Our strategy incorporates 3-fold improvements: (1) local data augmentations, (2) a hierarchically balanced architecture, and (3) a hybrid contrastive-restorative loss function. We evaluate our method on CT and MRI data and demonstrate that our new approach particularly beneficial for fine-tuning with limited annotated data and consistently outperforms the baseline counterpart in linear evaluation settings.
DG-TTA: Out-of-domain medical image segmentation through Domain Generalization and Test-Time Adaptation
Dec 22, 2023



Abstract:Applying pre-trained medical segmentation models on out-of-domain images often yields predictions of insufficient quality. Several strategies have been proposed to maintain model performance, such as finetuning or unsupervised- and source-free domain adaptation. These strategies set restrictive requirements for data availability. In this study, we propose to combine domain generalization and test-time adaptation to create a highly effective approach for reusing pre-trained models in unseen target domains. Domain-generalized pre-training on source data is used to obtain the best initial performance in the target domain. We introduce the MIND descriptor previously used in image registration tasks as a further technique to achieve generalization and present superior performance for small-scale datasets compared to existing approaches. At test-time, high-quality segmentation for every single unseen scan is ensured by optimizing the model weights for consistency given different image augmentations. That way, our method enables separate use of source and target data and thus removes current data availability barriers. Moreover, the presented method is highly modular as it does not require specific model architectures or prior knowledge of involved domains and labels. We demonstrate this by integrating it into the nnUNet, which is currently the most popular and accurate framework for medical image segmentation. We employ multiple datasets covering abdominal, cardiac, and lumbar spine scans and compose several out-of-domain scenarios in this study. We demonstrate that our method, combined with pre-trained whole-body CT models, can effectively segment MR images with high accuracy in all of the aforementioned scenarios. Open-source code can be found here: https://github.com/multimodallearning/DG-TTA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge