Jianqiang Li
Unveiling Scaling Behaviors in Molecular Language Models: Effects of Model Size, Data, and Representation
Jan 30, 2026Abstract:Molecular generative models, often employing GPT-style language modeling on molecular string representations, have shown promising capabilities when scaled to large datasets and model sizes. However, it remains unclear and subject to debate whether these models adhere to predictable scaling laws under fixed computational budgets, which is a crucial understanding for optimally allocating resources between model size, data volume, and molecular representation. In this study, we systematically investigate the scaling behavior of molecular language models across both pretraining and downstream tasks. We train 300 models and conduct over 10,000 experiments, rigorously controlling compute budgets while independently varying model size, number of training tokens, and molecular representation. Our results demonstrate clear scaling laws in molecular models for both pretraining and downstream transfer, reveal the substantial impact of molecular representation on performance, and explain previously observed inconsistencies in scaling behavior for molecular generation. Additionally, we publicly release the largest library of molecular language models to date to facilitate future research and development. Code and models are available at https://github.com/SZU-ADDG/MLM-Scaling.
From Tokens to Blocks: A Block-Diffusion Perspective on Molecular Generation
Jan 29, 2026Abstract:Drug discovery can be viewed as a combinatorial search over an immense chemical space, motivating the development of deep generative models for de novo molecular design. Among these, GPT-based molecular language models (MLM) have shown strong molecular design performance by learning chemical syntax and semantics from large-scale data. However, existing MLMs face two fundamental limitations: they inadequately capture the graph-structured nature of molecules when formulated as next-token prediction problems, and they typically lack explicit mechanisms for target-aware generation. Here, we propose SoftMol, a unified framework that co-designs molecular representation, model architecture, and search strategy for target-aware molecular generation. SoftMol introduces soft fragments, a rule-free block representation of SMILES that enables diffusion-native modeling, and develops SoftBD, the first block-diffusion molecular language model that combines local bidirectional diffusion with autoregressive generation under molecular structural constraints. To favor generated molecules with high drug-likeness and synthetic accessibility, SoftBD is trained on a carefully curated dataset named ZINC-Curated. SoftMol further integrates a gated Monte Carlo tree search to assemble fragments in a target-aware manner. Experimental results show that, compared with current state-of-the-art models, SoftMol achieves 100% chemical validity, improves binding affinity by 9.7%, yields a 2-3x increase in molecular diversity, and delivers a 6.6x speedup in inference efficiency. Code is available at https://github.com/szu-aicourse/softmol
Toward Closed-loop Molecular Discovery via Language Model, Property Alignment and Strategic Search
Dec 18, 2025Abstract:Drug discovery is a time-consuming and expensive process, with traditional high-throughput and docking-based virtual screening hampered by low success rates and limited scalability. Recent advances in generative modelling, including autoregressive, diffusion, and flow-based approaches, have enabled de novo ligand design beyond the limits of enumerative screening. Yet these models often suffer from inadequate generalization, limited interpretability, and an overemphasis on binding affinity at the expense of key pharmacological properties, thereby restricting their translational utility. Here we present Trio, a molecular generation framework integrating fragment-based molecular language modeling, reinforcement learning, and Monte Carlo tree search, for effective and interpretable closed-loop targeted molecular design. Through the three key components, Trio enables context-aware fragment assembly, enforces physicochemical and synthetic feasibility, and guides a balanced search between the exploration of novel chemotypes and the exploitation of promising intermediates within protein binding pockets. Experimental results show that Trio reliably achieves chemically valid and pharmacologically enhanced ligands, outperforming state-of-the-art approaches with improved binding affinity (+7.85%), drug-likeness (+11.10%) and synthetic accessibility (+12.05%), while expanding molecular diversity more than fourfold. By combining generalization, plausibility, and interpretability, Trio establishes a closed-loop generative paradigm that redefines how chemical space can be navigated, offering a transformative foundation for the next era of AI-driven drug discovery.
Automating Conflict-Aware ACL Configurations with Natural Language Intents
Aug 25, 2025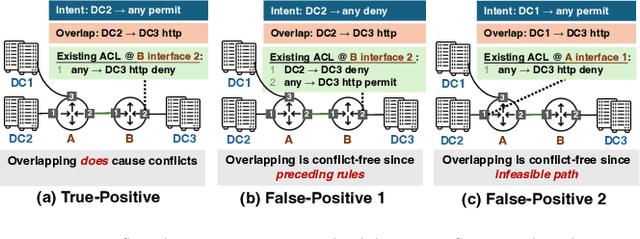
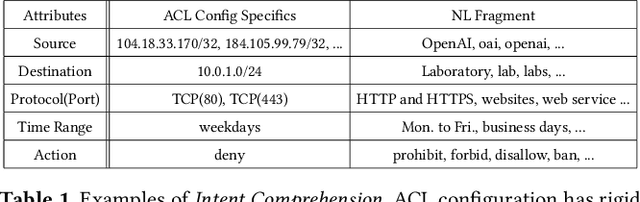
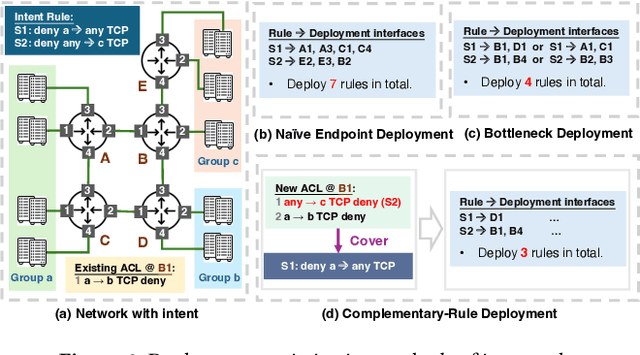
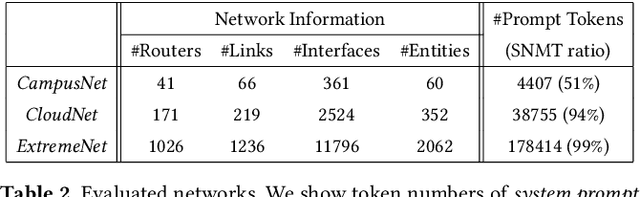
Abstract:ACL configuration is essential for managing network flow reachability, yet its complexity grows significantly with topologies and pre-existing rules. To carry out ACL configuration, the operator needs to (1) understand the new configuration policies or intents and translate them into concrete ACL rules, (2) check and resolve any conflicts between the new and existing rules, and (3) deploy them across the network. Existing systems rely heavily on manual efforts for these tasks, especially for the first two, which are tedious, error-prone, and impractical to scale. We propose Xumi to tackle this problem. Leveraging LLMs with domain knowledge of the target network, Xumi automatically and accurately translates the natural language intents into complete ACL rules to reduce operators' manual efforts. Xumi then detects all potential conflicts between new and existing rules and generates resolved intents for deployment with operators' guidance, and finally identifies the best deployment plan that minimizes the rule additions while satisfying all intents. Evaluation shows that Xumi accelerates the entire configuration pipeline by over 10x compared to current practices, addresses O(100) conflicting ACLs and reduces rule additions by ~40% in modern cloud network.
BiggerGait: Unlocking Gait Recognition with Layer-wise Representations from Large Vision Models
May 23, 2025



Abstract:Large vision models (LVM) based gait recognition has achieved impressive performance. However, existing LVM-based approaches may overemphasize gait priors while neglecting the intrinsic value of LVM itself, particularly the rich, distinct representations across its multi-layers. To adequately unlock LVM's potential, this work investigates the impact of layer-wise representations on downstream recognition tasks. Our analysis reveals that LVM's intermediate layers offer complementary properties across tasks, integrating them yields an impressive improvement even without rich well-designed gait priors. Building on this insight, we propose a simple and universal baseline for LVM-based gait recognition, termed BiggerGait. Comprehensive evaluations on CCPG, CAISA-B*, SUSTech1K, and CCGR\_MINI validate the superiority of BiggerGait across both within- and cross-domain tasks, establishing it as a simple yet practical baseline for gait representation learning. All the models and code will be publicly available.
ADNP-15: An Open-Source Histopathological Dataset for Neuritic Plaque Segmentation in Human Brain Whole Slide Images with Frequency Domain Image Enhancement for Stain Normalization
May 08, 2025Abstract:Alzheimer's Disease (AD) is a neurodegenerative disorder characterized by amyloid-beta plaques and tau neurofibrillary tangles, which serve as key histopathological features. The identification and segmentation of these lesions are crucial for understanding AD progression but remain challenging due to the lack of large-scale annotated datasets and the impact of staining variations on automated image analysis. Deep learning has emerged as a powerful tool for pathology image segmentation; however, model performance is significantly influenced by variations in staining characteristics, necessitating effective stain normalization and enhancement techniques. In this study, we address these challenges by introducing an open-source dataset (ADNP-15) of neuritic plaques (i.e., amyloid deposits combined with a crown of dystrophic tau-positive neurites) in human brain whole slide images. We establish a comprehensive benchmark by evaluating five widely adopted deep learning models across four stain normalization techniques, providing deeper insights into their influence on neuritic plaque segmentation. Additionally, we propose a novel image enhancement method that improves segmentation accuracy, particularly in complex tissue structures, by enhancing structural details and mitigating staining inconsistencies. Our experimental results demonstrate that this enhancement strategy significantly boosts model generalization and segmentation accuracy. All datasets and code are open-source, ensuring transparency and reproducibility while enabling further advancements in the field.
Automated Hybrid Reward Scheduling via Large Language Models for Robotic Skill Learning
May 05, 2025Abstract:Enabling a high-degree-of-freedom robot to learn specific skills is a challenging task due to the complexity of robotic dynamics. Reinforcement learning (RL) has emerged as a promising solution; however, addressing such problems requires the design of multiple reward functions to account for various constraints in robotic motion. Existing approaches typically sum all reward components indiscriminately to optimize the RL value function and policy. We argue that this uniform inclusion of all reward components in policy optimization is inefficient and limits the robot's learning performance. To address this, we propose an Automated Hybrid Reward Scheduling (AHRS) framework based on Large Language Models (LLMs). This paradigm dynamically adjusts the learning intensity of each reward component throughout the policy optimization process, enabling robots to acquire skills in a gradual and structured manner. Specifically, we design a multi-branch value network, where each branch corresponds to a distinct reward component. During policy optimization, each branch is assigned a weight that reflects its importance, and these weights are automatically computed based on rules designed by LLMs. The LLM generates a rule set in advance, derived from the task description, and during training, it selects a weight calculation rule from the library based on language prompts that evaluate the performance of each branch. Experimental results demonstrate that the AHRS method achieves an average 6.48% performance improvement across multiple high-degree-of-freedom robotic tasks.
Performance Estimation for Supervised Medical Image Segmentation Models on Unlabeled Data Using UniverSeg
Apr 22, 2025Abstract:The performance of medical image segmentation models is usually evaluated using metrics like the Dice score and Hausdorff distance, which compare predicted masks to ground truth annotations. However, when applying the model to unseen data, such as in clinical settings, it is often impractical to annotate all the data, making the model's performance uncertain. To address this challenge, we propose the Segmentation Performance Evaluator (SPE), a framework for estimating segmentation models' performance on unlabeled data. This framework is adaptable to various evaluation metrics and model architectures. Experiments on six publicly available datasets across six evaluation metrics including pixel-based metrics such as Dice score and distance-based metrics like HD95, demonstrated the versatility and effectiveness of our approach, achieving a high correlation (0.956$\pm$0.046) and low MAE (0.025$\pm$0.019) compare with real Dice score on the independent test set. These results highlight its ability to reliably estimate model performance without requiring annotations. The SPE framework integrates seamlessly into any model training process without adding training overhead, enabling performance estimation and facilitating the real-world application of medical image segmentation algorithms. The source code is publicly available
Comparative Analysis of Pre-trained Deep Learning Models and DINOv2 for Cushing's Syndrome Diagnosis in Facial Analysis
Jan 21, 2025



Abstract:Cushing's syndrome is a condition caused by excessive glucocorticoid secretion from the adrenal cortex, often manifesting with moon facies and plethora, making facial data crucial for diagnosis. Previous studies have used pre-trained convolutional neural networks (CNNs) for diagnosing Cushing's syndrome using frontal facial images. However, CNNs are better at capturing local features, while Cushing's syndrome often presents with global facial features. Transformer-based models like ViT and SWIN, which utilize self-attention mechanisms, can better capture long-range dependencies and global features. Recently, DINOv2, a foundation model based on visual Transformers, has gained interest. This study compares the performance of various pre-trained models, including CNNs, Transformer-based models, and DINOv2, in diagnosing Cushing's syndrome. We also analyze gender bias and the impact of freezing mechanisms on DINOv2. Our results show that Transformer-based models and DINOv2 outperformed CNNs, with ViT achieving the highest F1 score of 85.74%. Both the pre-trained model and DINOv2 had higher accuracy for female samples. DINOv2 also showed improved performance when freezing parameters. In conclusion, Transformer-based models and DINOv2 are effective for Cushing's syndrome classification.
Deep Learning-Based Feature Fusion for Emotion Analysis and Suicide Risk Differentiation in Chinese Psychological Support Hotlines
Jan 15, 2025



Abstract:Mental health is a critical global public health issue, and psychological support hotlines play a pivotal role in providing mental health assistance and identifying suicide risks at an early stage. However, the emotional expressions conveyed during these calls remain underexplored in current research. This study introduces a method that combines pitch acoustic features with deep learning-based features to analyze and understand emotions expressed during hotline interactions. Using data from China's largest psychological support hotline, our method achieved an F1-score of 79.13% for negative binary emotion classification.Additionally, the proposed approach was validated on an open dataset for multi-class emotion classification,where it demonstrated better performance compared to the state-of-the-art methods. To explore its clinical relevance, we applied the model to analysis the frequency of negative emotions and the rate of emotional change in the conversation, comparing 46 subjects with suicidal behavior to those without. While the suicidal group exhibited more frequent emotional changes than the non-suicidal group, the difference was not statistically significant.Importantly, our findings suggest that emotional fluctuation intensity and frequency could serve as novel features for psychological assessment scales and suicide risk prediction.The proposed method provides valuable insights into emotional dynamics and has the potential to advance early intervention and improve suicide prevention strategies through integration with clinical tools and assessments The source code is publicly available at https://github.com/Sco-field/Speechemotionrecognition/tree/main.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge